

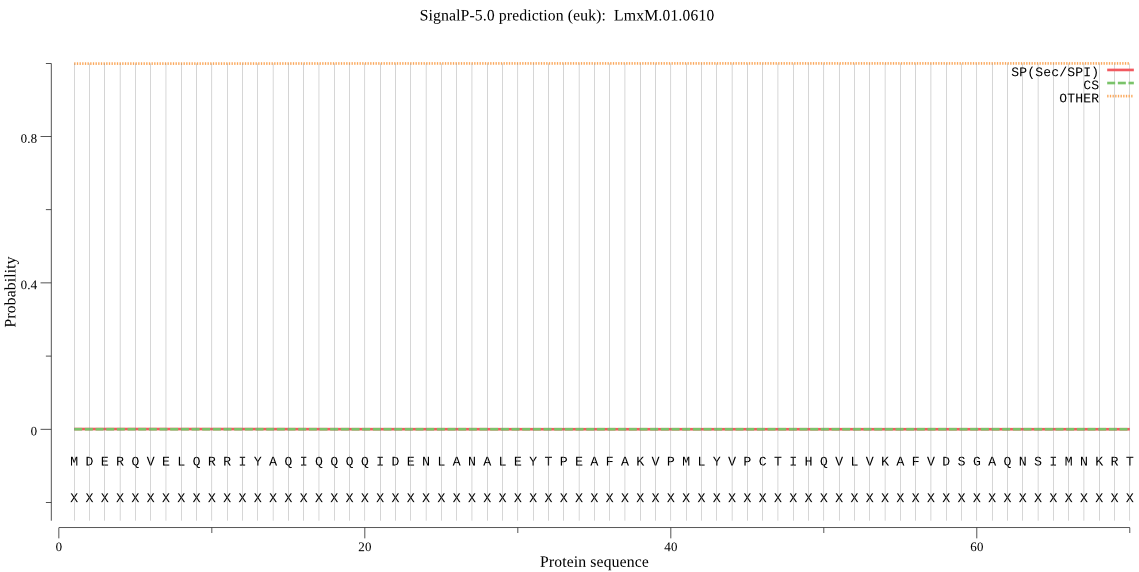
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.01.0610 | OTHER | 0.999951 | 0.000005 | 0.000044 |
MDERQVELQRRIYAQIQQQQIDENLANALEYTPEAFAKVPMLYVPCTIHQVLVKAFVDSG AQNSIMNKRTAERCGLMRLVDVRMRGVAVGVGRQEICGRIHMTPVNLAGMYIPFAFYVIE DQAMDLIIGLDQLRRHQMVIDLKHDCLTIGNINVPFLPENDLPALAGLDDDADEMHAPRH QDPAVTATTAPSPAAPVLSEGERQARMEGFMTFSGITDPKQAAELLEAADWDPNVAAALL FDT