

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.02.0710 | mTP | 0.364887 | 0.001169 | 0.633944 | CS pos: 62-63. CRW-QS. Pr: 0.3196 |
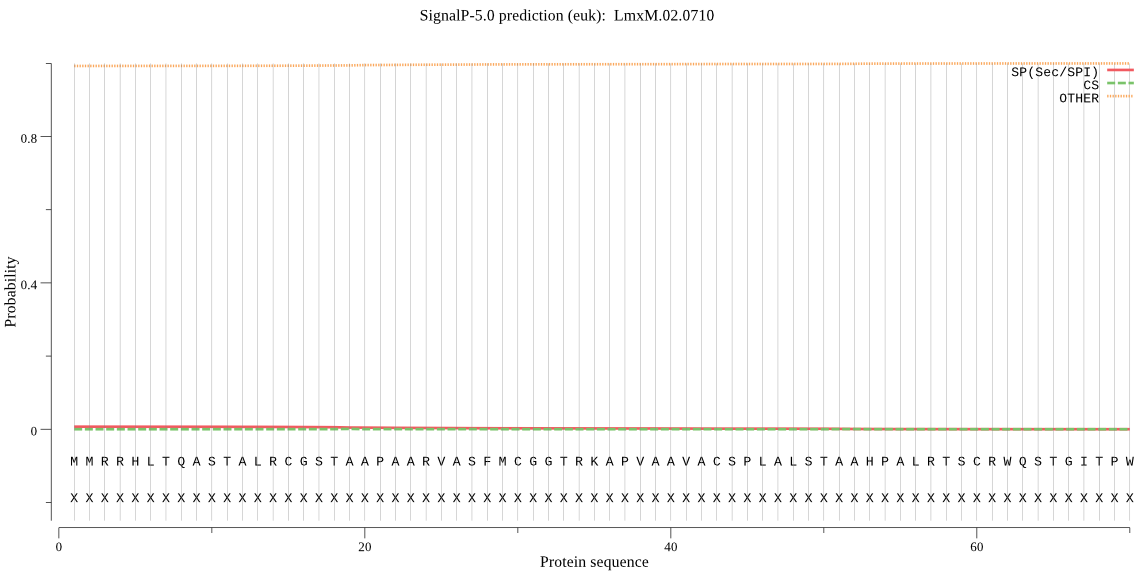
MMRRHLTQASTALRCGSTAAPAARVASFMCGGTRKAPVAAVACSPLALSTAAHPALRTSC RWQSTGITPWNSSGSSGGGGNGGQQQGRTWVNPNAVPPGEFLKKYARDLTEEARMGRLDP IVGREEVIRRTIQVVSRRTKNNPVLIGEPGVGKTAIVEGLAQRIVKGEVPESIKDRQVYA LDMGTLVAGAKFRGEFEERLKGVLKDTIESKGKVILFIDELHTLVGAGSSGDGSMDAANL LKPSLARGELHCIGATTLNEYRQHIEKDAALARRFQSVLVTEPTVEETISILRGIKEKYE AHHGCLIKDEALVYAAVNSHRYLSERRLPDKAIDLIDEAASRLRLQQESKPEALDLIERE LIRLKIEAEAVKKDKDEVGKEKLENLYEQIAQKQKEYDALEERWKKEKNLFHTIKQRTED LDVLRHHLEQAMSSGDFAKAGEIQHSQIPNLLKQIEKDKHLASSQNFMVHDSVTSKDIAE VIARATGIPVAQLMTSEREKLIHMDAELKKTIMGQDTAIESITNVVRISRAGLHSHKRPL GAFLFLGPTGVGKTEVCKSLAKFLFDDESFICRIDMSEYMERHSVHRLIGAPPGYVGYEE GGELTESVRRRPYQIVLFDEFEKAHPSVSNILLQVLDEGHLTDSHGRRVDFKNTIIILTS NIGADAIARLPEGEPSISAMPTVMEQVRNRLAPEFINRLDDIIMFNRLSRTDIRRIVEIL FAQVQKMLTEHNITLEGSTEVYDWLSVNGYSTVYGARPLKRLVQSELLNQLALMLLDGRI REGEHVKLSVRDSHVVVIANHALTPEKGPTDERLLDQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.02.0710 | 535 S | AGLHSHKRP | 0.995 | unsp | LmxM.02.0710 | 535 S | AGLHSHKRP | 0.995 | unsp | LmxM.02.0710 | 535 S | AGLHSHKRP | 0.995 | unsp | LmxM.02.0710 | 584 S | MERHSVHRL | 0.995 | unsp | LmxM.02.0710 | 709 S | FNRLSRTDI | 0.996 | unsp | LmxM.02.0710 | 789 S | HVKLSVRDS | 0.997 | unsp | LmxM.02.0710 | 172 S | EVPESIKDR | 0.997 | unsp | LmxM.02.0710 | 324 S | HRYLSERRL | 0.997 | unsp |