

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.08.0450 | OTHER | 0.914671 | 0.081530 | 0.003799 |
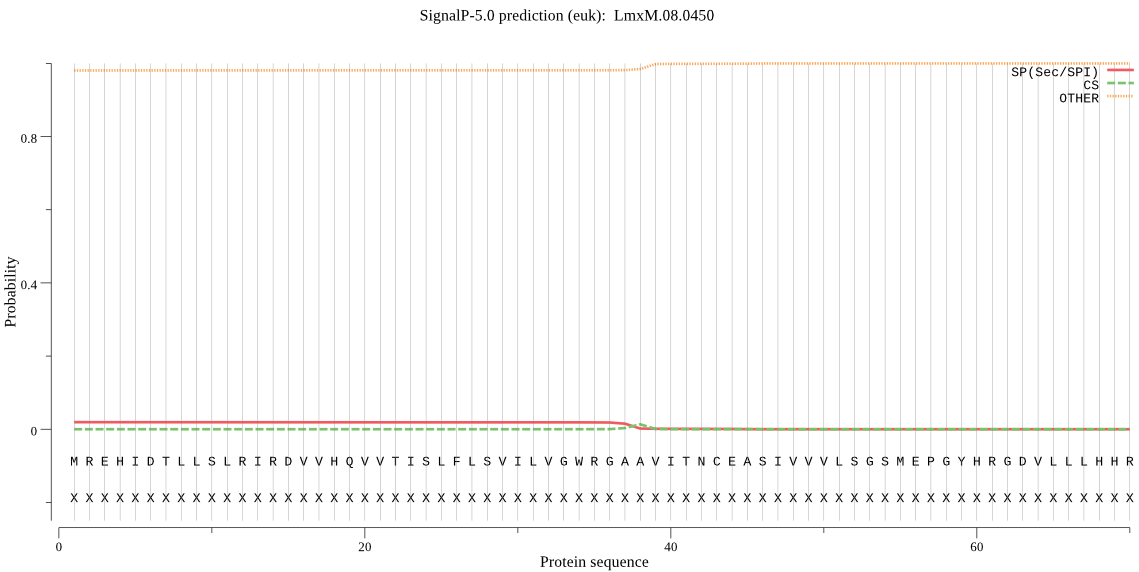
MREHIDTLLSLRIRDVVHQVVTISLFLSVILVGWRGAAVITNCEASIVVVLSGSMEPGYH RGDVLLLHHRPEYPVEVGDIIVYTLPGQEIPIVHRVHRIHERAEDHKRLYLTKGDNNMND DRFLFHGGREWVEQDMIIGKTFAYVPRIGYLTIMFNESKVIKYVALALLGFFMLTSTQDL