

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
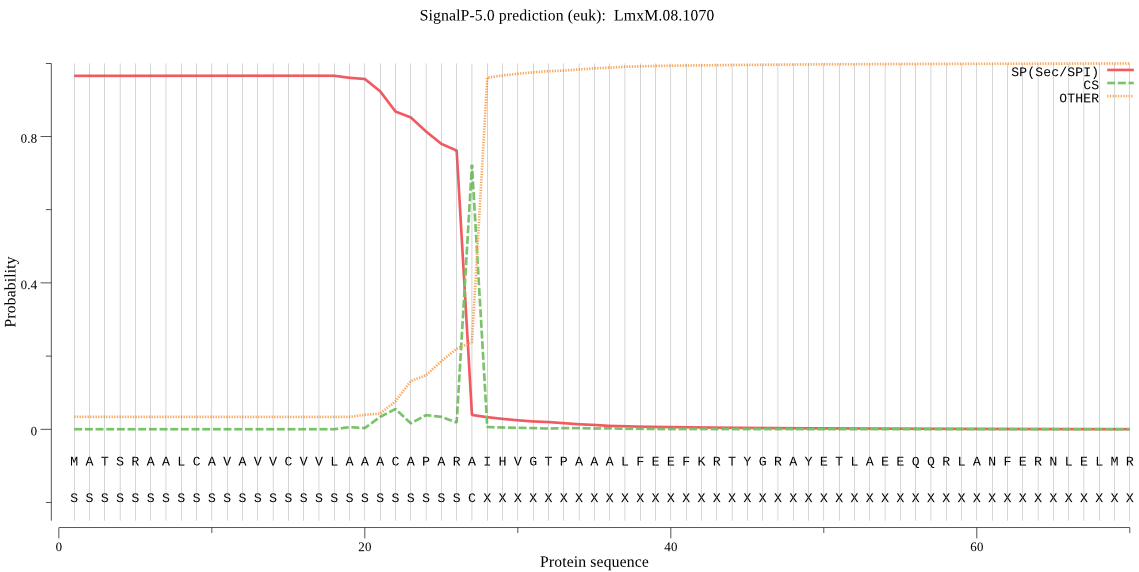
| LmxM.08.1070 | SP | 0.000923 | 0.996098 | 0.002979 | CS pos: 27-28. ARA-IH. Pr: 0.7165 |
MATSRAALCAVAVVCVVLAAACAPARAIHVGTPAAALFEEFKRTYGRAYETLAEEQQRLA NFERNLELMREHQARNPHAQFGITKFFDLSEAEFAARYLNGAAYFAAAKRHAAQHYRKAR ADLSAVPDAVDWREKGAVTPVKDQGACGSCWAFSAVGNIEGQWYLAGHELVSLSEQQLVS CDDMNDGCDGGLMLQAFDWLLQNTNGHLHTEDSYPYVSGNGYVPECSNSSELVVGAQIDG HVLIGSSEKAMAAWLAKNGPIAIALDASSFMSYKSGVLTACIGKQLNHGVLLVGYDMTGE VPYWVIKNSWGGDWGEQGYVRVVMGVNACLLSEYPVSAHVRESAAPGTSTSSETPAPRPV VVEQVICFDKNCRRGCRKTLIKVNECHKNGGGGASMIKCSPQKVTMCTYSNEFCVGGGLC FETHDGKCSPYFFGSIMNTCHYT
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmxM.08.1070 | 272 S | SSFMSYKSG | 0.993 | unsp |