

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
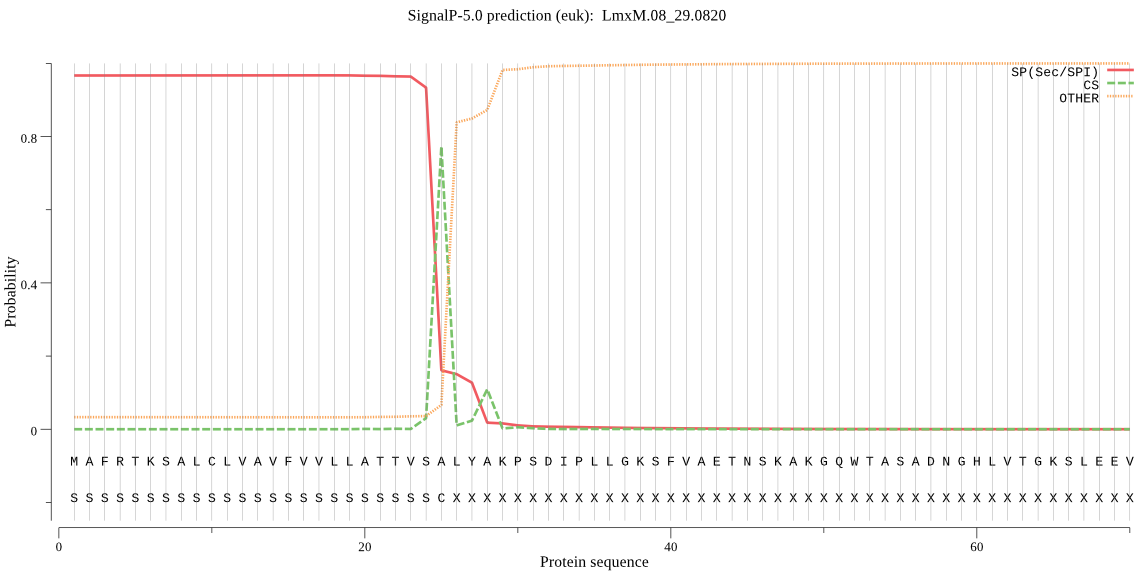
| LmxM.08_29.0820 | SP | 0.017374 | 0.982519 | 0.000107 | CS pos: 25-26. VSA-LY. Pr: 0.7223 |
MAFRTKSALCLVAVFVVLLATTVSALYAKPSDIPLLGKSFVAETNSKAKGQWTASADNGH LVTGKSLEEVRKLMGVTSMSTEAVPPRNFSVEEMQQDLPESFDASEKWPMCVTIGEIRDQ SNCGSCWAIAAVEAMSDRYCTMSGIPDRRISTTNLLSCCFICGFGCYGGIPAMAWLWWVW VGVTTELCQPYPFGPCSHHGNSSKYPPCPNTIYNTPKCNTTCDNVEMELVKYKGVSSYSI KGERELMVELMNNGPLEVAMQVYADFVAYKSGVYKHVSGDHLGGHAVKLVGWGVKDGIPY WKIANSWNTDWGDKGYFLIQRGNDECGIESSGVAGKPGEE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.08_29.0820 | 239 S | VSSYSIKGE | 0.993 | unsp | LmxM.08_29.0820 | 239 S | VSSYSIKGE | 0.993 | unsp | LmxM.08_29.0820 | 239 S | VSSYSIKGE | 0.993 | unsp | LmxM.08_29.0820 | 90 S | PRNFSVEEM | 0.996 | unsp | LmxM.08_29.0820 | 151 S | DRRISTTNL | 0.996 | unsp |