

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
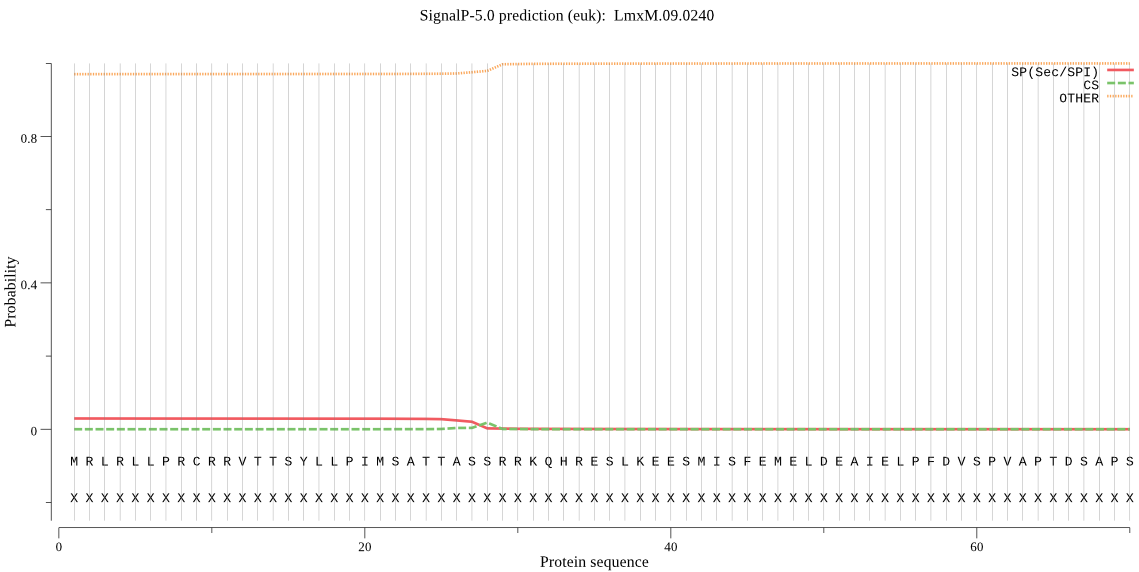
| LmxM.09.0240 | OTHER | 0.761247 | 0.058928 | 0.179825 |
MRLRLLPRCRRVTTSYLLPIMSATTASSRRKQHRESLKEESMISFEMELDEAIELPFDVS PVAPTDSAPSPPFGRKKIAMPASRTASEKRVAWSNGSTASLGLAVCGNVNELPNTAATAD GVTSPDVVPPNAAAKTYSAAAPSRMVTWKDTSASQRRAEAQASRQDDFNPMILCPLQNLG NTCYFNAGVQLLVNCPALVYAVRNSPFARRAQSNIPLTAHAVPMPPAAAPTATTKLCLKG GAATHALFQEFSALLARMEMGCAAGERAISPMCALDSLAQAYPQFEGRSQQDAAEMMTSL LASLEEEGGQYVEVTQLLHSFEEDAAALRQGVTSSASSPVGPELMGRVRTSATSSLLEAE AAPSASSHRRDYSEPYNIEEYSQPSLLSSTSEDTFLPGPRSAHFFAVLRLMERVNRDNEL LERRVKQRQGVAYTSPFKPPRLHFNPLLDGFRGYTLSEVECHNCRAVSRVVSAFNGLLLD VPTAKQRRRYALAHPNVLRRVDFDGELRQVKKPFRMTWWNPLSLCVAAWRQLKRFFQDPF PYPLWLDECLDIHFEPEVLRGCNKYRCESCDTTTDATKSETLLALPEYLLIHMKRFEAGR FFNSKKSDSVFFPTSWRPLTNTQARQSPINTDDAPPALPEYLDLRHYLHSSVASFAEPTP PCLWSADNEPHQQRRASRDDASSPTSSIPTTYTLDGIVNHHGGYDGGHYTVFLYKATEER QAWVYISDDEVDQAEEVVATDTEYVLLYRRQPLVQAAPESDDGEQLRRKACYYLSLSSPS CPAANTAAATTAGPHAKKLHSRCPFSSSSADGAASAPPDAATAATEPRVYISRMWLQRVA FMHEPGPIVNRLCYCRPEDQGRVSMYRTIFPATVKVPAEVPHVHGPPVSWFYVPITQKDY DIFYNAFGGNAAVTASEYESLRAMQDKFCAAIDLAERQRGRSARPAPRS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.09.0240 | 87 S | SRTASEKRV | 0.995 | unsp | LmxM.09.0240 | 87 S | SRTASEKRV | 0.995 | unsp | LmxM.09.0240 | 87 S | SRTASEKRV | 0.995 | unsp | LmxM.09.0240 | 154 S | DTSASQRRA | 0.996 | unsp | LmxM.09.0240 | 270 S | ERAISPMCA | 0.993 | unsp | LmxM.09.0240 | 303 S | SLLASLEEE | 0.995 | unsp | LmxM.09.0240 | 320 S | QLLHSFEED | 0.997 | unsp | LmxM.09.0240 | 338 S | SSASSPVGP | 0.997 | unsp | LmxM.09.0240 | 351 S | RVRTSATSS | 0.991 | unsp | LmxM.09.0240 | 367 S | PSASSHRRD | 0.993 | unsp | LmxM.09.0240 | 627 S | QARQSPINT | 0.998 | unsp | LmxM.09.0240 | 677 S | QRRASRDDA | 0.998 | unsp | LmxM.09.0240 | 683 S | DDASSPTSS | 0.996 | unsp | LmxM.09.0240 | 864 S | QGRVSMYRT | 0.997 | unsp | LmxM.09.0240 | 942 S | QRGRSARPA | 0.997 | unsp | LmxM.09.0240 | 36 S | QHRESLKEE | 0.998 | unsp | LmxM.09.0240 | 70 S | DSAPSPPFG | 0.995 | unsp |