

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
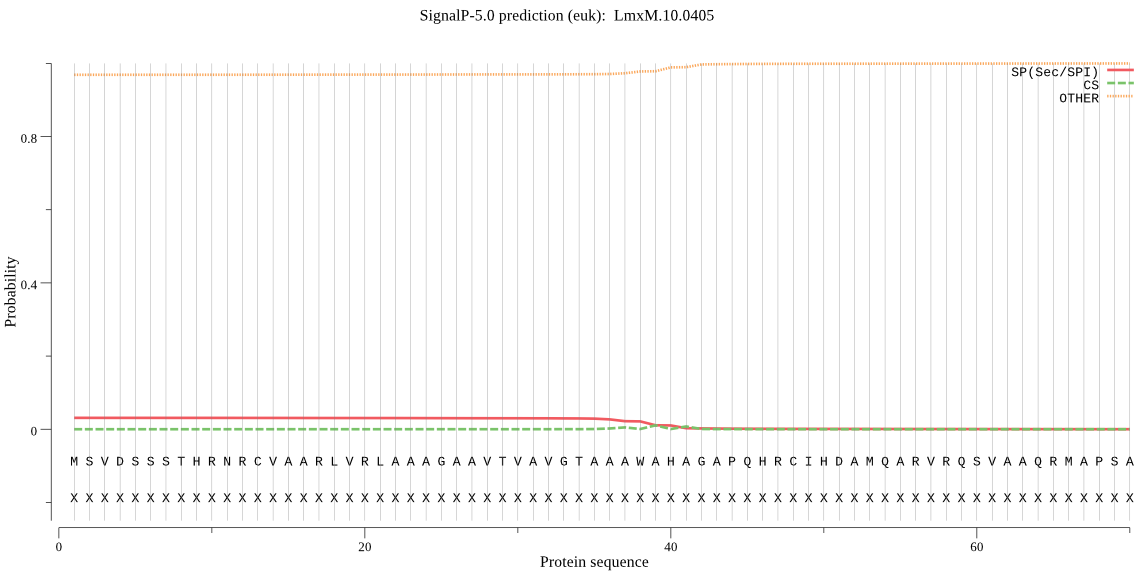
| LmxM.10.0405 | OTHER | 0.940968 | 0.032255 | 0.026777 |
MSVDSSSTHRNRCVAARLVRLAAAGAAVTVAVGTAAAWAHAGAPQHRCIHDAMQARVRQS VAAQRMAPSAVSAVGLPHVTLDAADTAAGADPSTGTPRNVVRAANWGALRIAVSAEDLTD PAYHCARVGQRISARDGRFAVCTAEDILTDEKRDILVKHLVPQALQLHRERLKVRQVQGK WKVTDMAADVCSYFKVPPAHVTGGVSNTDFVLYVASVPSEESVLAWAMTCQVFPDGHPAV GVINIPAANIASRYDQLVTRVVAHEMAHALGFSGTFFEAVGIVQEVPGIRGKTFTVAVIT SSTAVAKAREQYGCNSLEYLEIEDQGGAGSAGSHIKMRNAKDELMAPAASAGYYTALTMA VFQDLGFYQADFSKAEEMPWGRNVRCAFLSEKCMAKNVTKWPAMFCNESAATIRCPTDRL RVGTCGITAYNTSLATYWQYFTNASLGGYSPFLDYCPFVVGYRNGSCNQDASTTPDLLAA FNVFSEAARCIDGAFTPKNRTAADGYYTALCANVKCDTATRTYSVQVRGSNGYANCTPGL RVKLSSVSDAFEKGGYVTCPPYVEVCQGNVKAAKDFAGDTDSSSSADDAADKEAMQRWSD RMAALATATTLLLGMVLSLMALLVVRLLLTSSPWCCCRLGGLPT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.10.0405 | 114 S | RIAVSAEDL | 0.991 | unsp | LmxM.10.0405 | 114 S | RIAVSAEDL | 0.991 | unsp | LmxM.10.0405 | 114 S | RIAVSAEDL | 0.991 | unsp | LmxM.10.0405 | 133 S | GQRISARDG | 0.997 | unsp | LmxM.10.0405 | 546 S | VKLSSVSDA | 0.997 | unsp | LmxM.10.0405 | 584 S | TDSSSSADD | 0.991 | unsp | LmxM.10.0405 | 585 S | DSSSSADDA | 0.997 | unsp | LmxM.10.0405 | 7 S | VDSSSTHRN | 0.992 | unsp | LmxM.10.0405 | 96 T | PSTGTPRNV | 0.991 | unsp |