

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
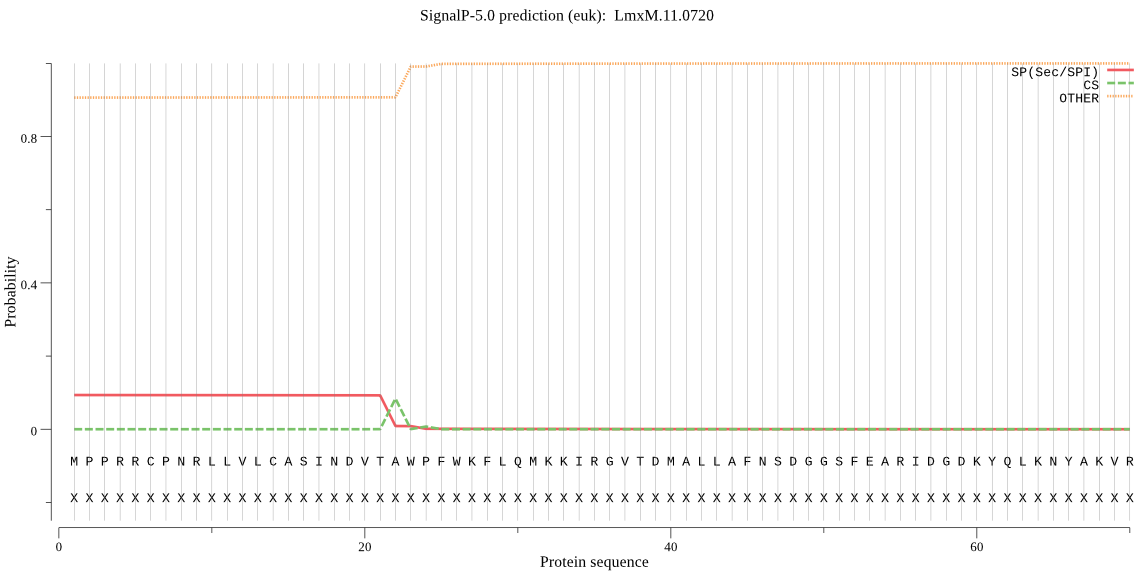
| LmxM.11.0720 | SP | 0.450414 | 0.543484 | 0.006103 | CS pos: 22-23. VTA-WP. Pr: 0.9666 |
MPPRRCPNRLLVLCASINDVTAWPFWKFLQMKKIRGVTDMALLAFNSDGGSFEARIDGDK YQLKNYAKVRGYQHDMFESFVHRWHDPGRSYFVYGGHGMGDYVELEQNRVSLQVHELADV FGTRIFEAVLFDACFMANLDCAYHLRHNTRYIGACEGYMWEPDTALDYHVFNTHNASAMS RFKDPLHILRVIQADYCSKAPRGDFTIIDTTHIAELRQYVQEHVMQRVYDRATFYSLPQR ERLQQIAEAAIQASISQFGHPGGDTNVMNGVGSGTGRPRTASLSPEMLALSAAGRPTRRQ RMLQAIQFEHSLYPSEVDDKQLLDLKSYLTDMLREEQQQKAWEAAALGPHRRIAAGRRRV RSDALEHGGSSGIAAASCASSSFAVWKAPPSRALFVDEHGRLPAASSAGCALSTNGGPVA ATDTQEEATALCPAALASTPAMAAAPPPSLSTSYKGSAQEGLNLFHQVVVSQIPPKAASI YASQLGGLSFTVHEYSAMSRPAEPWLVGSKRLLKRRAKQFLKSGELSEVVMESPKASVPV TGAALVAAVEKATATAAAVTGASAGKPQHATSVPALPLSPRTRMLSTDQRLGFASSSNGT DSDSSLSLSLSAPLFTSSTDACNSNMKAVILDSPQRPGSYTSLPDSKQRTSAC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.11.0720 | 533 S | VVMESPKAS | 0.995 | unsp | LmxM.11.0720 | 533 S | VVMESPKAS | 0.995 | unsp | LmxM.11.0720 | 533 S | VVMESPKAS | 0.995 | unsp | LmxM.11.0720 | 639 S | QRPGSYTSL | 0.993 | unsp | LmxM.11.0720 | 646 S | SLPDSKQRT | 0.993 | unsp | LmxM.11.0720 | 284 S | TASLSPEML | 0.996 | unsp | LmxM.11.0720 | 453 S | SLSTSYKGS | 0.994 | unsp |