-
-
-
Computed_GO_Function_IDs: GO:0008233
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>LmxM.15.0580
MHSMRQRSCDIVASTVSALLVTAVLTASSTILRDLVLRPRPLASDVRVSSLSPLYHVRSP
TLLSPEIPDAAIYYRPGTGSDGRRSAEKMLPPNIKIVDTVDRPEDSEEDRQRKTWQRQRL
PKGFELGVRTIDRVALYLSGSVDFSPCWDWNTKAIYISFIARFPSTSASQNDVVLLDAVL
RPVSLPAKLARLAKLRRQERSAALPGSGNGRATVAARASPAATQMLTDEERQQLAAFEQT
LRNVDHDHPSVYIDHIDKRIVLNDSFKYFVDAFDDTSLGGNTVEVVLRYQVMSYSGWAPV
REDVLGHQVKVKMPENAVLWKAHQR
- Download Fasta
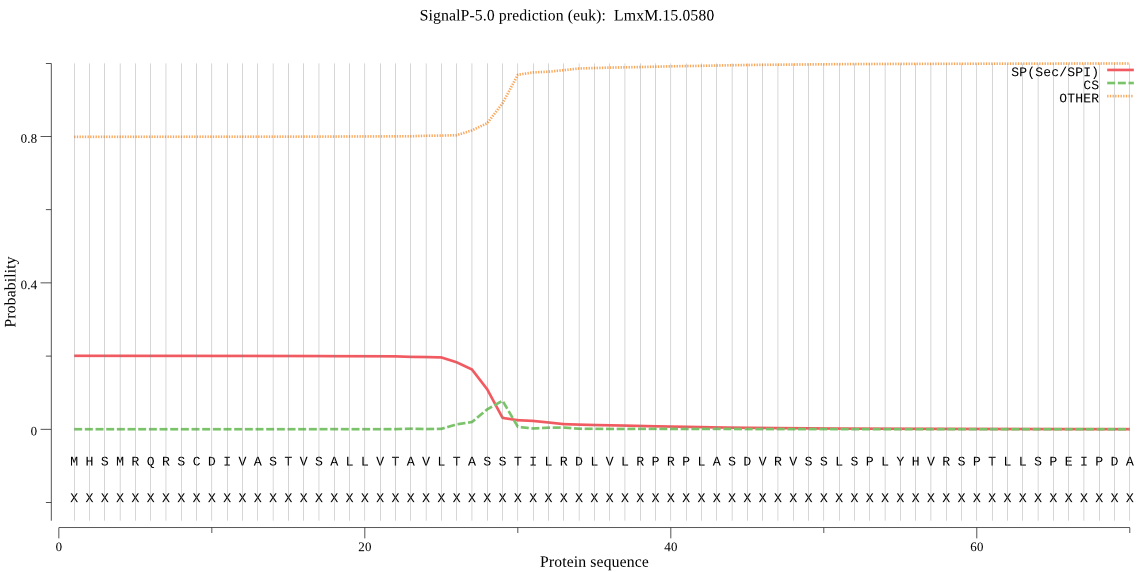
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/637
Sequence name : 637
Sequence length : 325
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.827
CoefTot : 1.497
ChDiff : 6
ZoneTo : 33
KR : 3
DE : 1
CleavSite : 43
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.947 2.129 0.371 0.651
MesoH : -0.458 0.481 -0.230 0.172
MuHd_075 : 20.621 11.200 5.808 3.839
MuHd_095 : 24.948 20.700 7.645 6.567
MuHd_100 : 26.411 17.608 7.382 5.606
MuHd_105 : 25.259 13.154 6.594 4.605
Hmax_075 : 12.600 23.100 2.861 5.017
Hmax_095 : 15.137 21.087 4.740 7.040
Hmax_100 : 16.000 22.100 5.028 5.930
Hmax_105 : 14.000 16.275 3.972 4.445
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.8823 0.1177
DFMC : 0.7747 0.2253
-
Fasta :-
>LmxM.15.0580
ATGCACTCGATGCGACAGCGCTCCTGCGACATCGTTGCCTCGACCGTGTCGGCCCTACTC
GTCACGGCAGTCCTAACGGCCAGCTCGACGATTTTGCGAGACCTGGTGCTGCGCCCGCGT
CCGCTCGCCTCGGACGTGCGTGTGTCGAGCCTCTCGCCGCTGTACCACGTGCGCAGCCCT
ACGCTGCTGTCTCCAGAGATCCCGGATGCGGCCATCTACTACCGCCCCGGCACCGGCAGC
GATGGCCGCAGAAGCGCGGAAAAAATGCTCCCGCCGAATATTAAGATCGTGGACACGGTG
GATCGTCCTGAGGACAGCGAGGAAGATCGCCAGCGCAAGACATGGCAGCGGCAGCGCTTA
CCGAAAGGCTTCGAGCTCGGCGTACGCACCATCGATCGCGTCGCGCTGTACCTCTCTGGC
TCGGTGGACTTCTCCCCCTGTTGGGACTGGAACACGAAAGCTATCTACATCTCCTTCATT
GCCCGCTTTCCGTCGACGTCGGCATCTCAGAATGACGTCGTCCTGCTCGATGCGGTGCTG
CGCCCGGTGTCACTGCCAGCGAAGCTCGCGCGGTTGGCGAAGTTGCGTCGCCAAGAGCGG
AGCGCGGCGCTACCGGGAAGCGGCAACGGGAGAGCGACGGTGGCGGCCAGGGCCTCTCCC
GCCGCCACGCAGATGTTGACGGACGAAGAGCGGCAGCAGCTCGCTGCCTTCGAGCAGACT
CTGCGCAATGTCGATCACGATCACCCCTCCGTGTACATTGACCACATCGACAAGCGGATC
GTGCTGAACGACTCATTCAAGTACTTTGTGGACGCCTTTGACGACACCTCTCTCGGCGGC
AACACGGTTGAGGTGGTACTGCGCTACCAGGTGATGAGCTACTCCGGCTGGGCGCCCGTG
CGGGAGGATGTGCTTGGGCACCAGGTGAAAGTAAAGATGCCAGAGAACGCTGTGCTGTGG
AAGGCGCATCAGCGGTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
LmxM.15.0580 | 85 S | DGRRSAEKM | 0.995 | unsp | LmxM.15.0580 | 106 S | RPEDSEEDR | 0.996 | unsp |


