

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
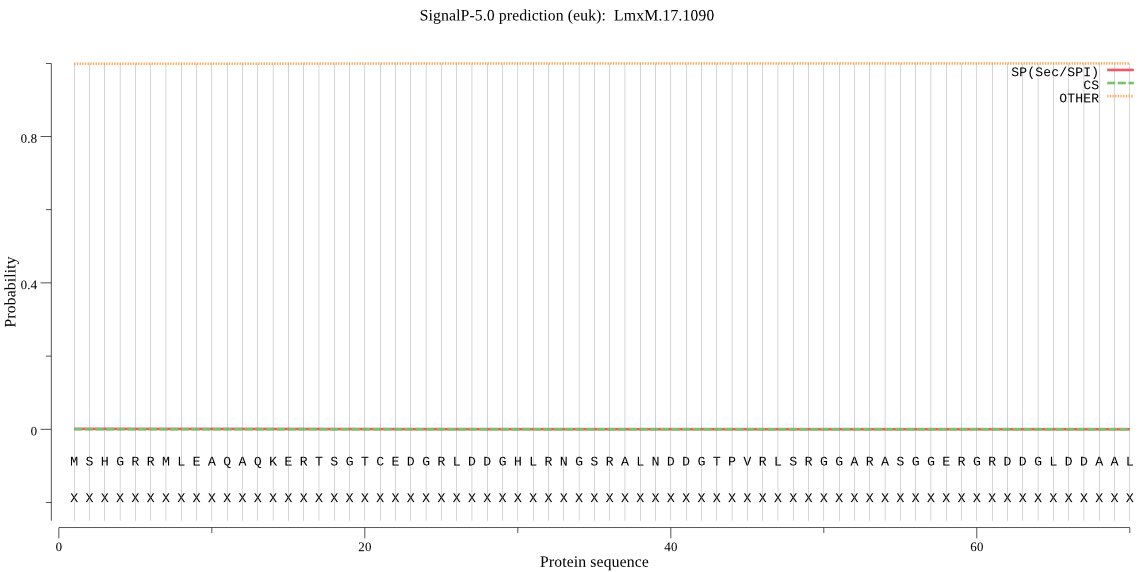
| LmxM.17.1090 | OTHER | 0.999975 | 0.000002 | 0.000023 |
MSHGRRMLEAQAQKERTSGTCEDGRLDDGHLRNGSRALNDDGTPVRLSRGGARASGGERG RDDGLDDAALVSPPPRPSVQLGFDRAGDGSGSASSVATAAGSALGATLAETANRQQRISG IGPSPAAPFSCAPLAVPRRLQSSDVAEGVRVSAARRIDVDAEEGESDDNVRKTRGTLAEA AGRGSPRRKGCGGRGDSFAGGDSAQRRMSSEARSRSRAGGSGHSSASGLRYPVSRVNALA STATLSAAPHDLPSPLNRSRTGSRSPLPMTAVQKTPASTSLARQPRAAPLPAPVTAAVLR PSNTESRASPTPDTGTATAATHAPRQPPPLRQESSSRSAGGPGFTTAMTSSAKTPPLQTL MSPASPPLRAGVSPRMLPLHPTRADASSASMFLSRAASASTVTTTVATTPSSSQAQLPPP VGSQVSPYELQQHQPYPIPPTPSSTAVAVMPSRSAVPHDMVDLAGDASGEDRAATETEPH TLLRSTKPLSQKSEPAVATAGSEWQSAPPSPLPTSTELLARPRLPLLPEATVPYDEVGVT PLTNSADRRVYPFMRDFSGFHNTGNDCYGCSLLTMLLRSEVFRRALLGSPLVCAMRRYEA LLTGKAERRVLWTSGYVNTAAGTRAKKAAARKRTRAAGFSIDDGDDGGQTEEASVAGACG LPDTTPRVPAETLAHSPYVWRPTADVQHTLDVLDTFSLSELEDVLEVDLETQRVPATLHG ALAALARAQRWREHMTTLVNAPRMQAAERQRLMETKIYHDNDRDFDGQVYTYGIRLNAVA NLFDGEFFLGDQEDAHELFVSVMAKLETEAVKLQHRCDEVLERRSSVTLTEASSAEDEVD EEVESEEGRPNNERGTRFAEKRLLHGAHAPFAPSLSPSDVPAAANPSEQPRLSVSTTAGE VWINTLVQTRLLNIIRCRTGECHHEIVTDEVCVNLSVHIPEERPTSSPPAPPLPPRPCFP AQPPPLLPTPIFPSSSVAATATAPLPAVDGGGGRCCSLANLLHESMAYEALNEYRCDCCG SRTSQFQGGCFYTRPPPLLVLQLKRFATQFVNGTIIIQRNGRRVAVEDKLVIHALPSAGE WHAGQRRQDRQHRRGPDPVESIEVTEAVEDARESAECYFYYDPDKKRFSAALQEVCDVSG GGGLAGQDGLKDERQHSTGDDLHTTLAAAAPPSSLVRAIRCLYRLRSCVLHLGQSLHYGH YVSDFAVDGEEDDEEDGEDEETARGRRTAEQEAGVRPTSEPTLSSFSAVRRRWRRANDER VEVLSDEVVQARRAGCSDTYLLLYEKVAEERVWCPAEAVLPAPVYRSSEGGKT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.17.1090 | 119 S | QQRISGIGP | 0.994 | unsp | LmxM.17.1090 | 119 S | QQRISGIGP | 0.994 | unsp | LmxM.17.1090 | 119 S | QQRISGIGP | 0.994 | unsp | LmxM.17.1090 | 185 S | AGRGSPRRK | 0.996 | unsp | LmxM.17.1090 | 209 S | QRRMSSEAR | 0.997 | unsp | LmxM.17.1090 | 214 S | SEARSRSRA | 0.993 | unsp | LmxM.17.1090 | 263 S | SRTGSRSPL | 0.99 | unsp | LmxM.17.1090 | 302 S | VLRPSNTES | 0.996 | unsp | LmxM.17.1090 | 309 S | ESRASPTPD | 0.998 | unsp | LmxM.17.1090 | 334 S | LRQESSSRS | 0.993 | unsp | LmxM.17.1090 | 336 S | QESSSRSAG | 0.99 | unsp | LmxM.17.1090 | 490 S | TKPLSQKSE | 0.997 | unsp | LmxM.17.1090 | 640 S | AAGFSIDDG | 0.994 | unsp | LmxM.17.1090 | 826 S | ERRSSVTLT | 0.997 | unsp | LmxM.17.1090 | 834 S | TEASSAEDE | 0.997 | unsp | LmxM.17.1090 | 876 S | APSLSPSDV | 0.995 | unsp | LmxM.17.1090 | 893 S | QPRLSVSTT | 0.994 | unsp | LmxM.17.1090 | 1114 S | DARESAECY | 0.995 | unsp | LmxM.17.1090 | 1157 S | ERQHSTGDD | 0.998 | unsp | LmxM.17.1090 | 18 S | KERTSGTCE | 0.995 | unsp | LmxM.17.1090 | 48 S | PVRLSRGGA | 0.996 | unsp |