

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.18.0360 | SP | 0.004138 | 0.995749 | 0.000113 | CS pos: 25-26. AAA-AS. Pr: 0.2730 |
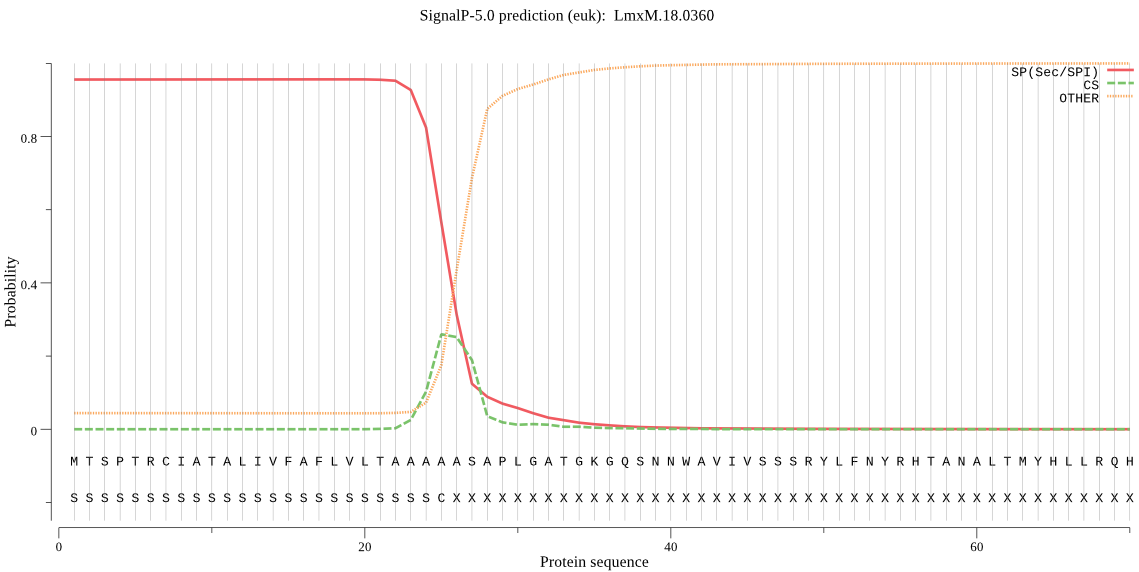
MTSPTRCIATALIVFAFLVLTAAAAASAPLGATGKGQSNNWAVIVSSSRYLFNYRHTANA LTMYHLLRQHGIDDDHILLFLSDSFACDPRNVYPAEIFSQPPGAHDADGRASMNLYGCSA QVDYAGSDVDVRRFLSVLQGRYDENTPPTRRLLSDNTSNIIIYVAGHGAKSYFKFQDTEF LSSSDISETLTMMHQQRRYGRVVFLADTCHAIALCEHVEAPNVVCLAASDAESESYSCQY DEQLGTHMVSFWMNEMYLLLNGTSCSNPLTRRIGDDAVSVLHQSWYNFNYHPYRVEASRN RSKPAHRDAVNDPTALREWIVADFVCSQVSAAVPVDVRYDLE