

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.18.0450 | SP | 0.000153 | 0.999842 | 0.000005 | CS pos: 29-30. VHA-ST. Pr: 0.8358 |
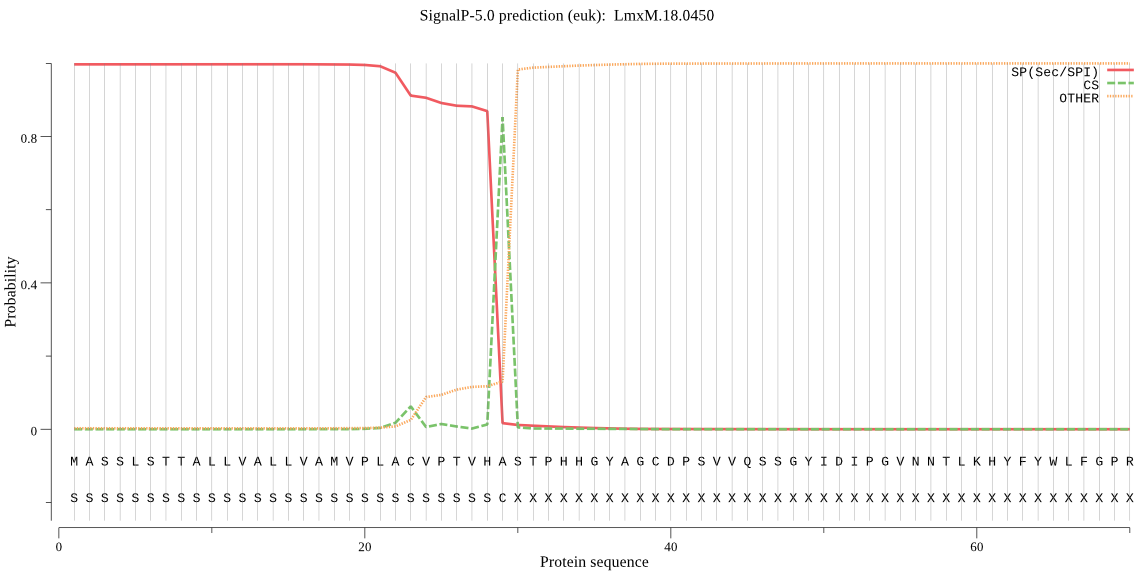
MASSLSTTALLVALLVAMVPLACVPTVHASTPHHGYAGCDPSVVQSSGYIDIPGVNNTLK HYFYWLFGPRKWSNDGREPPVIMWMTGGPGCSSTMALLTELGPCMMNETSGELYYNTYGW NDEAYLLFVDQPTGVGYSYGDKFNYVHNQSEVAEDMYNFLQLFARRFTSPSIIGTNDFYI IGESYAGHYVPAVSYRIVMGNERGDGLHINLKGIAVGNGITDPYTQLPFNAETAYYWCKE KLGFPCVTEKAYEEMISLLPACLEKTKKCNEGPDDSDVSCSVSTALWAQYVDYYYATGRN SYDIRKQCIGDLCYPMQNTIDFYHKPSVRASLGVSAEAQWSTCNSEVSVLFERDYMRNFN FTFPLMLDLGIRVLIYAGDMDFICNWLGNEAWVKALQWFGTDGFNSAPNVEFAVSGRWAG LERSYGGLSFVRIYDAGHMVPMDQPEVALFMVRRFLHGQNLA