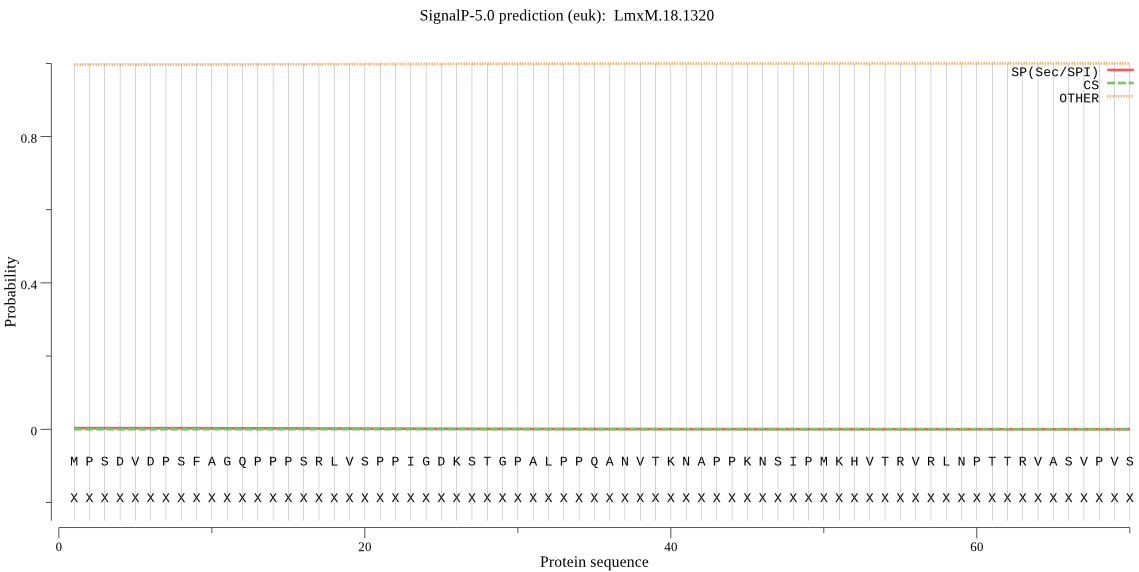
Fasta :-
MPSDVDPSFAGQPPPSRLVSPPIGDKSTGPALPPQANVTKNAPPKNSIPMKHVTRVRLNP
TTRVASVPVSPVPPLAGMPPGSGFRDSFVPRAATTAPTSAPQTHSDDHQVVEAMQAAYQK
TEVHLKAIEAKLNLLHKEIQRTASPATSRELPTEALGNNSTAESVLGSLVPIATTGAPGS
KAAANHHLQPIPSAMATPLPPVPLTSASTSTVVAKPKTSAATSAVVESSATAAAATPATA
KANRQHSPLDLHFVDHADILDSPNASIGSDSSSNDSSDSSSDAGHTLPNAARHSAGKPAF
SPAQTPAAATDHSEPSNRTAAQVRQRVDLKDVSHANACSSAVAASDRRAMRFREGGERSR
TAHSFPASLSSATPPADRGSTWGTGERGEVVAVGREFRQFSSQASTLTDAKVCFSLGESG
NHAVDNAQAVDRLGGVSRRQANVVKHVRERFFAPIPRFDDVLPRNKAGLFNKRDIEKLLL
PGAPCSWLCCASLAISYVSRVPTPVEKVLRANRMGVHYISLPTITLAELFDVVSDYIHVC
YHRTAVHKSSCTEDEFEDDAEASRCLSEDELRQLAHIHCEMATFDKEVLDPEFGEDMQGM
GEHPPIMNPTQLRKELLLHLGEEKSMYIFYYNPYVLEQAQLRLRSNQCDTEEEAAAVMAT
ARFSANVAGLFGILLDFNSVSHEVKLLTPYLSAEEHPLHFKDDEGEEGVHARHDVRRFAS
TFSNLVVEEQLVSLHALYESVQQSDKYLGLSRGYVRVFMSESFPPKVPSMFPLFVLDGSS
AGGLVTSVLNVKIAPHVLGLAMLHHLSVTFLLTESARRKQSSRNLLGKTSVCDVKLRGIP
LTKVCQQLRLPLSMIVCESNKGSIEMAYGWYHVFLQQLQIDQDVCIGLVLPSRRDGAADG
QPNITDDRFVHHLQLMIESKSVMLISFDMNIALNVRIDSRSDPAHFAIVIGVDEERGVVR
LADVNVKRFRKTWHIPISRLYNAVMGYGYIVASRSRRIIKALEGKHFHENALQCARNFLP
PPTPAAYHRFEYPPCPYPLTVLADAVERLGFAGTNVEHFLNFCGFHISYFLSPNMPLEGA
AFVIQNYSHYALDDAVSVATTHYDFYEGNLPPKEEAAEAGGTPTYPVRSEADLLESIQYA
IAEPDKRKLIVRYDVNVLQADETVWNGNHGNSFAFVVDYVASKKMVVLSDASPSSFYRSF
ACPLAVLFNAMCSWDSVSLRAHGTILLTTEVTQESMHEDTKGYDMAHALVHHPFKPMFSA
VCSCLALASTEMMMNIENPRLLSDAPISVEDEHRKYRRENNIFSTEDFLYALPSFSVCRW
RTQAVDIQDVGSVANRAFATLKLPLRIVDVLQEQGPDSRTDPKALLRACNGISGLQTITL
VTYDAGVLHGAPGLSAGLLNRVRLFDDENKEATVQPTVPGAPRRSVGSVQLLEGDPCRWG
ACFERSAEELMRAIKGIYRVEEAMHEDADG