

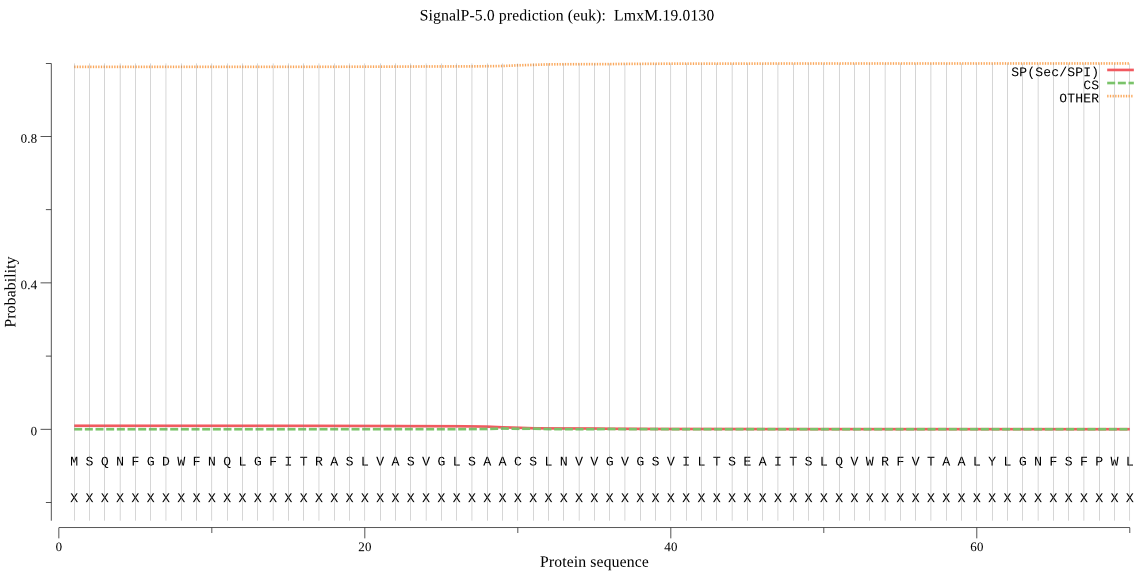
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.19.0130 | OTHER | 0.999622 | 0.000133 | 0.000246 |
MSQNFGDWFNQLGFITRASLVASVGLSAACSLNVVGVGSVILTSEAITSLQVWRFVTAAL YLGNFSFPWLMTVAMFVTYVKNNEESDFKGKTADMAYMFLLLVGVLSSAGLFFNVYVTSF SFLMALCWIFCKRHPEQELTLFGFSFRSAVFPWVLMALHLVMGQGLLADVFGIVAGHAYI FFKDVFPVSHNQRWLETPMWLRRQFPQPTHRVGSFGPEVHPYDPRFQAAWRGAAQQRSSG SHNWGRGQTLGSS