

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.19.1420 | SP | 0.007200 | 0.992310 | 0.000490 | CS pos: 22-23. CYG-SA. Pr: 0.3915 |
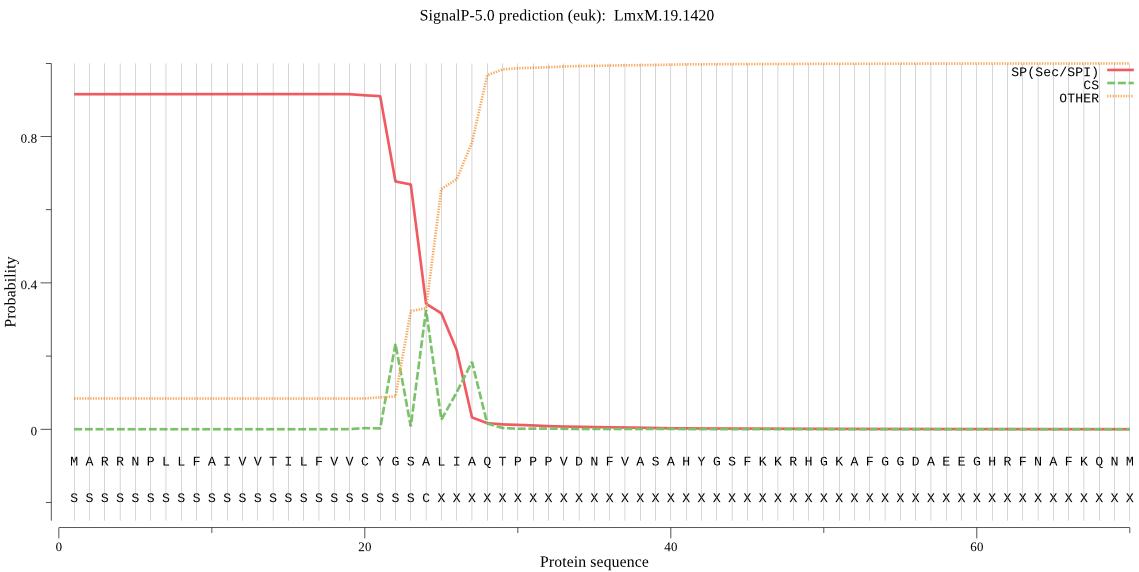
MARRNPLLFAIVVTILFVVCYGSALIAQTPPPVDNFVASAHYGSFKKRHGKAFGGDAEEG HRFNAFKQNMQTAYFLNTQNPHAHYDVSGKFADLTPQEFAKLYLNPDYYARHLKDHKEDV HVDDSAPSGVMSVDWRDKGAVTPVKNQGLCGSCWAFSAIGNIEGQWAASGHSLVSLSEQM LVSCDNIDEGCNGGLMDQAMNWIMQSHNGSVFTEASYPYTSGGGTRPPCHDEGEVGAKIT GFLSLPHDEERIAEWVEKRGPVAVAVDATTWQLYFGGVVSLCLAWSLNHGVLIVGFNKNA KPPYWIVKNSWGSSWGEKGYIRLAMGSNQCMLKNYPVSATVESPHTPHVPTTTA