

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
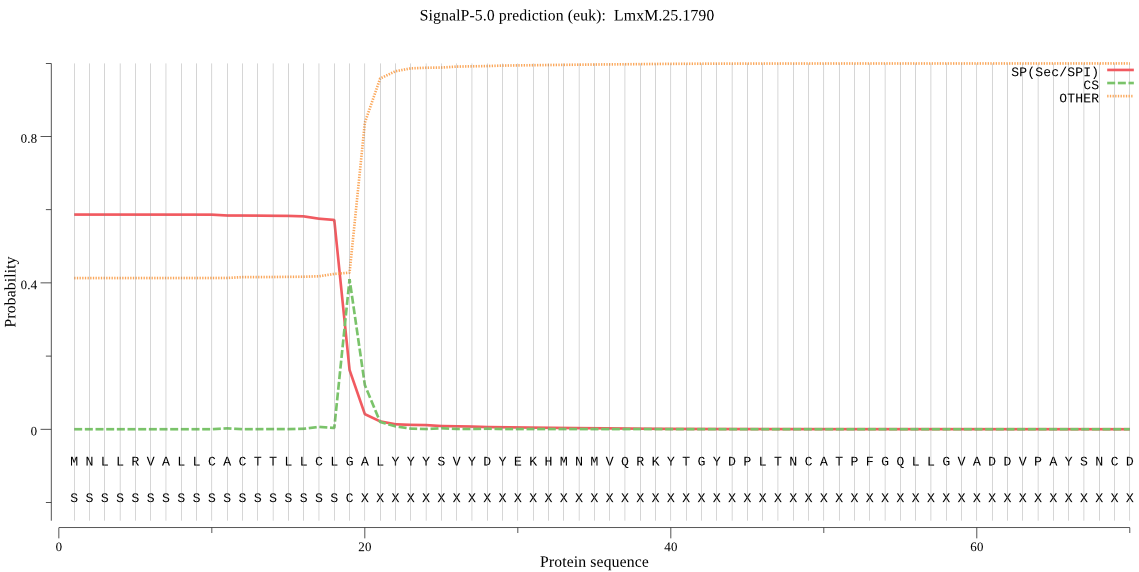
| LmxM.25.1790 | SP | 0.026575 | 0.973339 | 0.000086 | CS pos: 19-20. CLG-AL. Pr: 0.7078 |
MNLLRVALLCACTTLLCLGALYYYSVYDYEKHMNMVQRKYTGYDPLTNCATPFGQLLGVA DDVPAYSNCDIKFSSTYINYVNLMDPMDNGRRGDPSETRIIMTAYRYTSFDYGMRWLVWN RGVMPRLVENTNQLWKTVDYFNPARPEQTWSAEYITNYEEVTDVEERKFNAPRRADVIVY RMDKNTIPAGHIAVVVKVEDDVEAAGGPEKLKELKKMRLHPRHVYVAEQNWKNQPWGGHN YSRVLQFKWRTVSEKAHEGSYVDPDGLDIIGVVRVGKAMPLRTAPDPYQEALDMDNDGDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.25.1790 | 261 Y | HEGSYVDPD | 0.99 | unsp | LmxM.25.1790 | 261 Y | HEGSYVDPD | 0.99 | unsp | LmxM.25.1790 | 261 Y | HEGSYVDPD | 0.99 | unsp | LmxM.25.1790 | 253 S | WRTVSEKAH | 0.996 | unsp | LmxM.25.1790 | 260 S | AHEGSYVDP | 0.992 | unsp |