

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
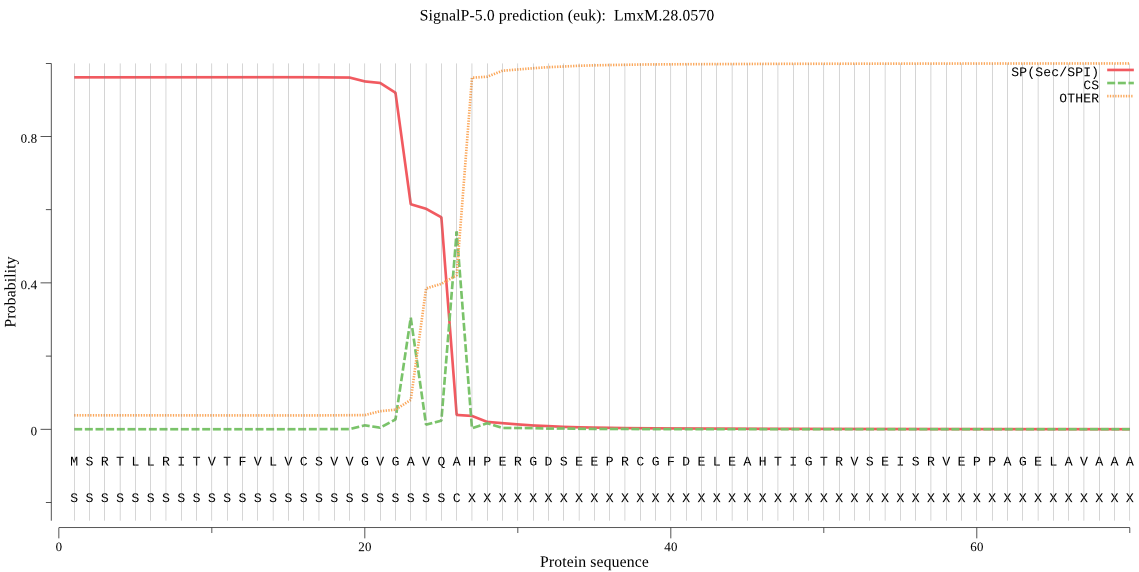
| LmxM.28.0570 | SP | 0.003625 | 0.996243 | 0.000132 | CS pos: 26-27. VQA-HP. Pr: 0.5068 |
MSRTLLRITVTFVLVCSVVGVGAVQAHPERGDSEEPRCGFDELEAHTIGTRVSEISRVEP PAGELAVAAAATGAWQPIRIAVFTDDISNSSQHCTASRQSRPSFRGDRVTCSAADVLTNR KKQALLELLIPSAVELHKERLNVQRENGNIVVNPFINKHPICGQFSIPKDHMKIGVKDAD FVLYMSAGPTSDNTIAWALKCQSFEDGRPSVGVANISPKYIAADPKIVRVIAHEVLHALG FTHSVFKEQNMLVMASFRGKSPSPVIRSEKVVAMARQHYGCQTQAFMELEDEGEKGSVSS HWKRRNAKDELMASVSGVGIYSALTIAAMEDTGYYKGNYDKAESMAYGHLAGCKLKDDRC VIKGTSQNNNMFCDAPDAPSSCTSDRRGVGKCIFTYYDSNLPTQFQYFENPRLGGSDPLM DFCPFVGDADDTKCTAKTNALKGSVYGIMSRCVDTPVGFSVGGSAAQQHGICAEVQCGSS TYGVKVRGASAFQDCLPGKTYNLSTLSPSFSKGYLVCPSYESVCAIKINASLYEEYSRFL TDHSVTGVRTSVTAVVAVLLVVLFMA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.28.0570 | 100 S | ASRQSRPSF | 0.991 | unsp | LmxM.28.0570 | 100 S | ASRQSRPSF | 0.991 | unsp | LmxM.28.0570 | 100 S | ASRQSRPSF | 0.991 | unsp | LmxM.28.0570 | 103 S | QSRPSFRGD | 0.998 | unsp | LmxM.28.0570 | 203 S | LKCQSFEDG | 0.995 | unsp | LmxM.28.0570 | 384 S | SSCTSDRRG | 0.992 | unsp | LmxM.28.0570 | 33 S | ERGDSEEPR | 0.995 | unsp | LmxM.28.0570 | 56 S | VSEISRVEP | 0.992 | unsp |