

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.29.1380 | OTHER | 0.999158 | 0.000428 | 0.000415 |
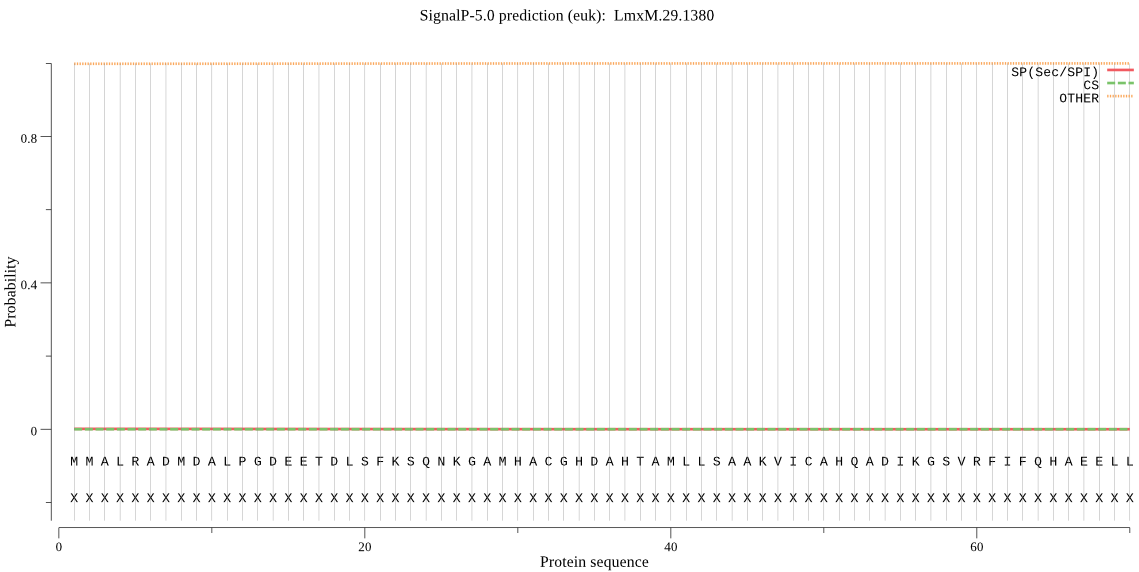
MMALRADMDALPGDEETDLSFKSQNKGAMHACGHDAHTAMLLSAAKVICAHQADIKGSVR FIFQHAEELLPGGAKDLVAAGVMEGVKSIFGMHCSPRYEAGTVGLMPGINTSYTDFFKLV ICGRGGHASTPQLLVDPVPITAEIVMAIQTITARKIDPRVVPVVSITTMTTGPNESHNVI PNEVKLMGTVRSRDKAVREQVPRDIERLASNIAAAHGAAATLDFTFGYDCCDNDPEVTAQ VRCIAERILGSANIIDPKVPLYGGEDFSAYQLKKPGCFLWLGTANKSQGICEMCHSTKFR VDETALPIGVSFHVGYVYDNVMA