

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
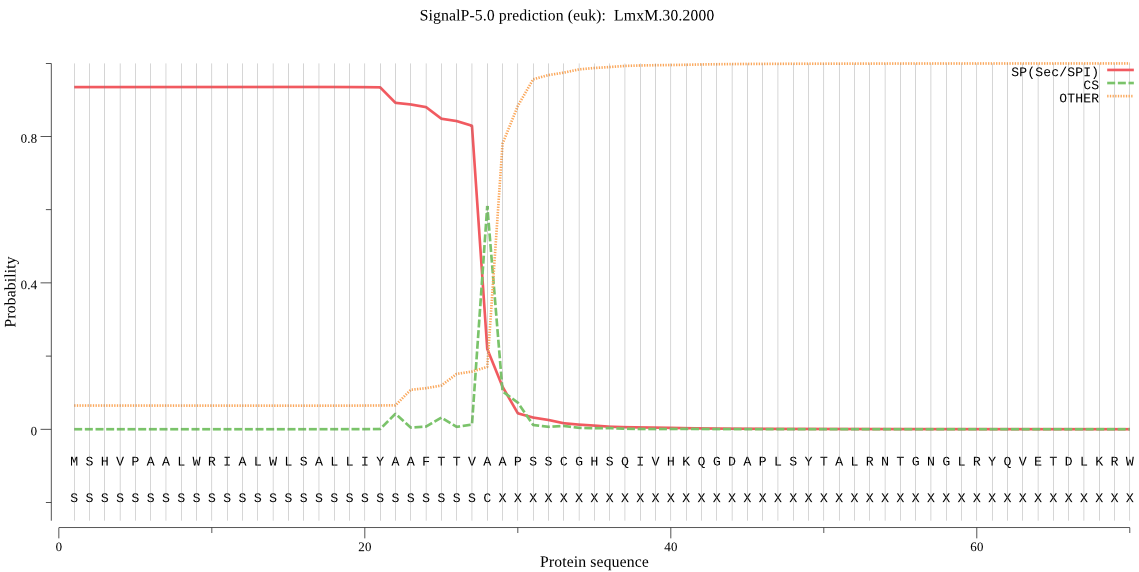
| LmxM.30.2000 | SP | 0.015773 | 0.983386 | 0.000841 | CS pos: 28-29. TVA-AP. Pr: 0.5534 |
MSHVPAALWRIALWLSALLIYAAFTTVAAPSSCGHSQIVHKQGDAPLSYTALRNTGNGLR YQVETDLKRWNGSATAEQRAVEPQATVTISSDREDSEWAPIRIAVFVPDLEDESKYCTAI GQMRPDSSGSLVPCTSNADLLTDAKRAVLINSILPQAVSMHAQRLKVRRLAASTSIVVNS MPGSYCGSFTVPTAHRTTGVPDADFIVYVAAGPTSTPKSFMAWALACQYYPNTATITSRP AVGAMYFNPRYLPTSVGETQEQIDRYGGNPSGSTNSLRRVAAHELLHALGFTSSIFKARG MFAIVPSLRGKRNVPVLNSSAVRAAAKAMYGISDTEVFYGAELEDQGEPGTSLSHWKRRA AKDELMAPVLSLARYSALSLAALEDMGFYKVDFRKAEPVALGATAGGKLFTVPCLTDGAS NTPTVFCDSLSSSVRSCTADRLSIGRCALTTYSSALPRYAQYFPDQPTLGGSLSHSDYCP VIQPLSNTACSGGSLGAMPGSITGDSARCFDADNLLVKFTLTQPGAVCAKVHCSSASRTY EVLVIGANSWLSCGAGGTGVTVQPAVSSPNVFVEGGFIACPPYDDVCYANPVAFAEVEAA ATTTTVSPSAATASTHGLVVSGLVAVAVLLGACIC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.30.2000 | 91 S | VTISSDRED | 0.994 | unsp | LmxM.30.2000 | 255 S | YLPTSVGET | 0.996 | unsp |