-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
-
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
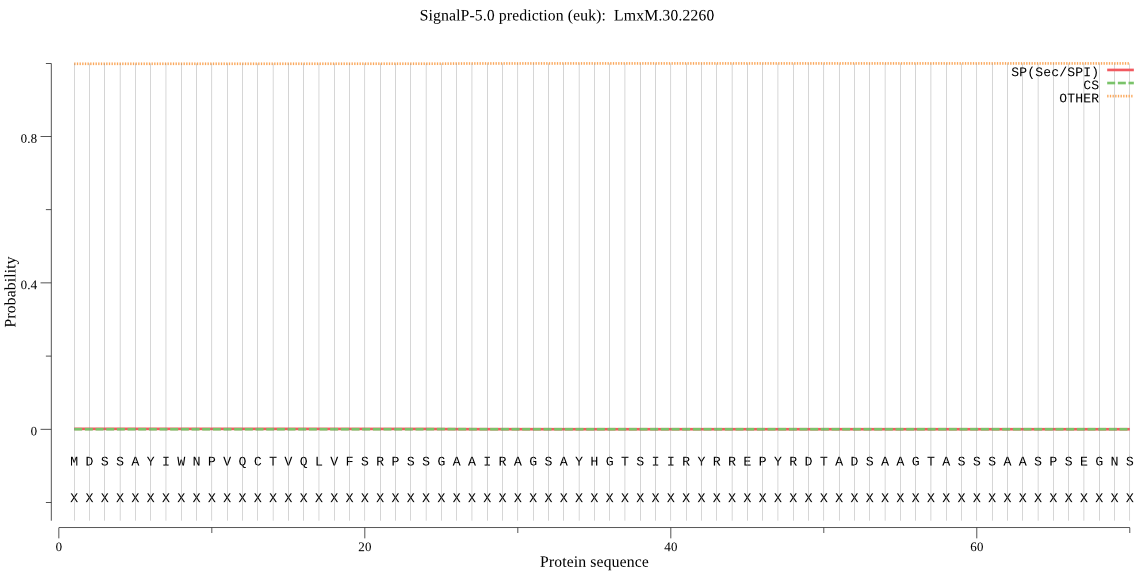
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
LmxM.30.2260 | OTHER | 0.995220 | 0.000281 | 0.004499 | |
No Results
-
Fasta :-
>LmxM.30.2260
MDSSAYIWNPVQCTVQLVFSRPSSGAAIRAGSAYHGTSIIRYRREPYRDTADSAAGTASS
SAASPSEGNSEYEDFLRHRRVIAEKNALHLHALTTAGGIYKDARVLDVQSYQVRCVSISA
DAGAAVLRVVSVEQEDAGAASAGSSTSGGAGKRKGSSKVWKKGGTDVAEHDAQTSGDSPT
PAAVDWGSKLILFEGAGSLPPFSEVDLTTQFIGRVQSFDCGGIYAAGGISSNPSTEDVPL
LTHFEVRFARCAFPAPDDPQYRLEWQLKSIQVPSSFRTILTNGEERGRKELAAQQAVQVS
FAPCGPLPAYVFSFACFPGIAEVESMSAAGDGLEVVEGSLDVPKFAGDIVSGLSDTSNLS
HTPVPVRVLARRQARIATATLERVLRLTIESVIALQHLFQCPLPLLQCEHLDVLLGPTMP
YISGMEHHCSIILNEAIYQPGRKTAAAGGGSAHSTAEVEQTELIVHELTHHWVGNALGLP
FAVKEGICQVIEQCVGDTLLGKPMRTYKADSSGSAKPESSSPSSSTAKPPKSTIRASEKG
HEFTGTSYQHALSAIKRLVAEHGFDRFVACLRQLMHVHVVVPAIAVEESGGIQVLRYVRS
GIATPPYLSTEQFLRNVESAL
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/504
Sequence name : 504
Sequence length : 621
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.135
CoefTot : -0.362
ChDiff : -9
ZoneTo : 44
KR : 5
DE : 1
CleavSite : 45
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.724 1.512 0.365 0.632
MesoH : 0.064 0.592 -0.162 0.338
MuHd_075 : 24.514 16.677 7.222 5.341
MuHd_095 : 23.969 19.941 5.805 7.065
MuHd_100 : 27.198 20.696 7.821 5.630
MuHd_105 : 31.353 23.237 8.558 6.473
Hmax_075 : 14.800 14.400 3.990 4.580
Hmax_095 : 8.200 14.700 3.359 5.819
Hmax_100 : 16.800 17.300 2.813 5.970
Hmax_105 : 16.300 17.100 3.205 5.180
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.2563 0.7437
DFMC : 0.2957 0.7043
This protein is probably imported in mitochondria.
f(Ser) = 0.1591 f(Arg) = 0.1136 CMi = 0.69444
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>LmxM.30.2260
ATGGACTCATCGGCGTACATATGGAACCCTGTCCAGTGCACGGTGCAGCTCGTCTTCAGC
CGCCCCAGCAGCGGCGCGGCGATTCGAGCTGGCTCCGCGTACCACGGCACATCCATCATC
CGCTACCGGCGTGAGCCGTATCGTGACACAGCAGACAGCGCCGCGGGAACTGCTAGCTCT
TCTGCAGCTTCTCCCTCTGAAGGCAACTCCGAGTACGAAGACTTCCTGCGACACCGTCGC
GTGATAGCCGAGAAAAACGCCCTGCATCTTCACGCACTCACAACCGCTGGCGGTATATAC
AAGGACGCCAGGGTGCTCGATGTCCAGTCCTATCAGGTGAGGTGTGTCTCCATCTCTGCT
GACGCGGGCGCGGCAGTGCTCAGGGTTGTGTCCGTCGAGCAGGAGGATGCTGGAGCGGCA
TCAGCGGGCAGCAGCACCAGTGGTGGAGCCGGGAAGAGGAAGGGAAGCTCCAAGGTTTGG
AAAAAGGGCGGCACGGATGTCGCGGAACACGATGCACAGACATCTGGCGACTCGCCCACA
CCCGCCGCAGTGGACTGGGGATCGAAGCTGATCCTATTCGAAGGCGCCGGCAGCTTACCC
CCGTTCTCTGAGGTGGATCTAACAACTCAGTTTATTGGCAGAGTTCAGTCTTTCGACTGT
GGTGGAATCTACGCCGCCGGTGGGATCAGCAGTAACCCCAGCACCGAAGATGTACCTCTA
CTCACACATTTTGAGGTGCGCTTCGCCCGGTGCGCGTTTCCGGCCCCCGACGACCCGCAG
TACCGCCTCGAATGGCAGCTCAAGAGTATCCAGGTGCCTAGTAGCTTCCGCACAATCTTG
ACGAACGGAGAGGAGCGTGGCCGCAAAGAACTGGCGGCGCAGCAGGCGGTGCAGGTGAGC
TTTGCCCCGTGCGGCCCGCTACCCGCGTACGTCTTCTCGTTTGCGTGCTTTCCAGGCATT
GCTGAGGTGGAGAGCATGTCCGCAGCTGGCGATGGGCTGGAGGTGGTAGAGGGAAGCTTG
GACGTCCCAAAGTTTGCTGGTGACATCGTGAGCGGCCTCAGTGACACCAGTAACCTGTCC
CACACACCCGTTCCTGTGCGTGTACTGGCGCGACGACAGGCGCGAATCGCTACGGCAACC
CTCGAGCGCGTCTTACGCCTCACCATCGAGTCTGTGATAGCGCTCCAGCACCTCTTTCAG
TGTCCACTGCCGCTCCTTCAGTGCGAGCACCTGGACGTCCTCCTTGGTCCAACAATGCCG
TATATCTCCGGTATGGAGCACCATTGCTCGATTATTCTCAACGAGGCCATCTACCAGCCA
GGCAGGAAAACCGCCGCCGCCGGTGGAGGGAGTGCGCACAGTACCGCCGAGGTGGAACAA
ACAGAGCTCATTGTGCACGAGCTGACACACCACTGGGTCGGAAACGCCCTGGGGCTGCCG
TTTGCCGTGAAGGAGGGTATCTGCCAGGTGATCGAGCAGTGTGTCGGCGACACGCTACTA
GGCAAGCCCATGCGCACGTACAAGGCAGACAGCAGCGGTAGCGCAAAGCCCGAGAGCTCC
TCCCCTTCCTCCTCGACTGCGAAGCCGCCGAAGAGCACCATCCGGGCCTCTGAGAAAGGC
CACGAGTTCACAGGGACCTCGTACCAGCACGCACTCAGTGCCATTAAGCGGTTGGTCGCC
GAGCACGGGTTTGACCGCTTTGTCGCGTGCCTGCGACAGCTCATGCACGTGCACGTCGTT
GTTCCAGCCATCGCAGTCGAGGAGAGCGGCGGAATCCAGGTGCTGCGGTACGTCCGTTCT
GGCATCGCCACCCCACCGTACCTAAGTACGGAGCAGTTCCTGCGTAACGTGGAGAGCGCG
TTGTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
LmxM.30.2260 | 131 S | LRVVSVEQE | 0.992 | unsp | LmxM.30.2260 | 131 S | LRVVSVEQE | 0.992 | unsp | LmxM.30.2260 | 131 S | LRVVSVEQE | 0.992 | unsp | LmxM.30.2260 | 156 S | KRKGSSKVW | 0.995 | unsp | LmxM.30.2260 | 234 S | SSNPSTEDV | 0.997 | unsp | LmxM.30.2260 | 511 S | YKADSSGSA | 0.995 | unsp | LmxM.30.2260 | 521 S | PESSSPSSS | 0.993 | unsp | LmxM.30.2260 | 532 S | KPPKSTIRA | 0.991 | unsp | LmxM.30.2260 | 537 S | TIRASEKGH | 0.998 | unsp | LmxM.30.2260 | 64 S | SSAASPSEG | 0.997 | unsp | LmxM.30.2260 | 70 S | SEGNSEYED | 0.994 | unsp |


