

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
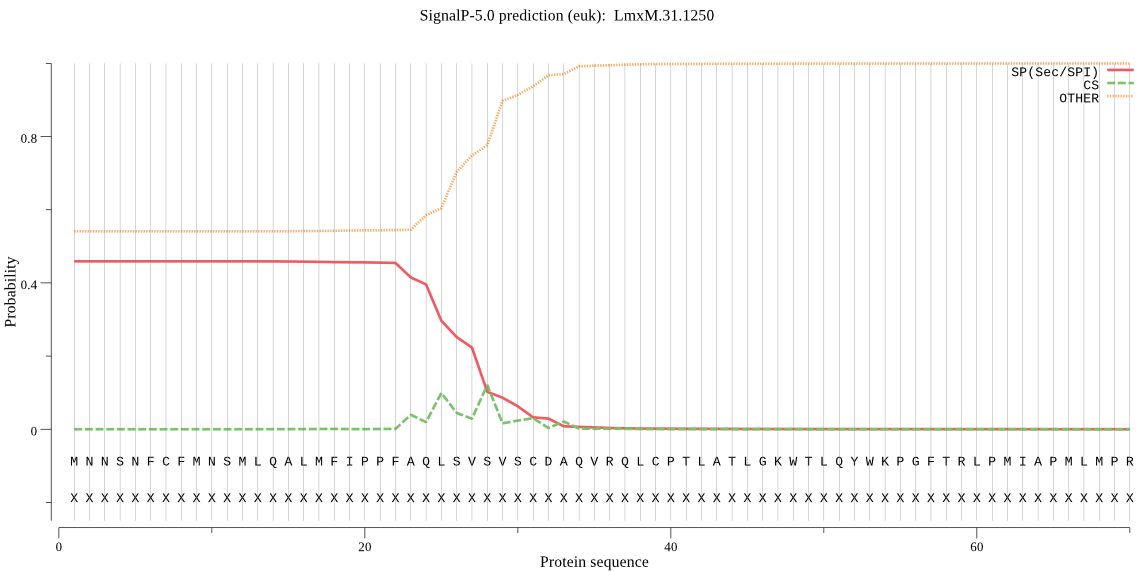
| LmxM.31.1250 | OTHER | 0.651372 | 0.340423 | 0.008205 |
MNNSNFCFMNSMLQALMFIPPFAQLSVSVSCDAQVRQLCPTLATLGKWTLQYWKPGFTRL PMIAPMLMPRMPAGAKSSNSGAPRSAVIPTSQRILDGSVQEDAQEFLQKLLERVYEELVS LEEAFQQADASEAATESDMGSNAPAVAASSASGDDRTDARDDATTSTFQKKGWTFVKGKE KLAVREYEDAQGQSKLMASIFGGTLESHLQGKQRQRNRVSVLIEKYYCLPVDVGFAPECT VEQALERTFMTERIYDSEHEQHLKKTLRLGHLPSILFLQLRRWAVTREGELVKLDNVVRI KRTLLIPRTICGDETLGNTERTYRLLSVVCHRGDAVSRGHYVTYLVHHAATPAVLKVQSS DPGNKDTAILRSPPDTATVILCNDANISVCPAKNMEKETMYFLVYQKTS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.31.1250 | 372 S | AILRSPPDT | 0.996 | unsp | LmxM.31.1250 | 372 S | AILRSPPDT | 0.996 | unsp | LmxM.31.1250 | 372 S | AILRSPPDT | 0.996 | unsp | LmxM.31.1250 | 120 S | EELVSLEEA | 0.993 | unsp | LmxM.31.1250 | 327 S | YRLLSVVCH | 0.993 | unsp |