

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.32.1800 | OTHER | 0.998990 | 0.000851 | 0.000159 |
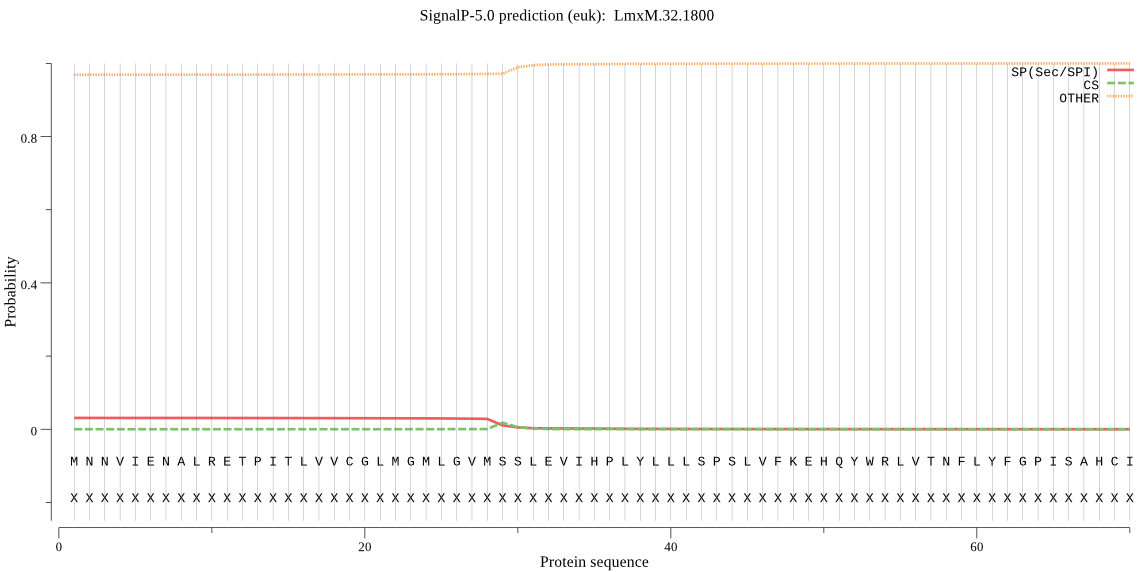
MNNVIENALRETPITLVVCGLMGMLGVMSSLEVIHPLYLLLSPSLVFKEHQYWRLVTNFL YFGPISAHCIMEIQWIYLVSSYLEAQYYHRRPLDYIFLLLIIGCSLLGLRFSSIVNVPYL SYMLGTCLTYIMSRLFNDMEVAIFFVVPVPMRLLPFVLLIMNTMVSGMTNEVLGNVLGHI LWYFLEVFPRITGQSPLRIQRLLERAFAAPVRQAT