

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
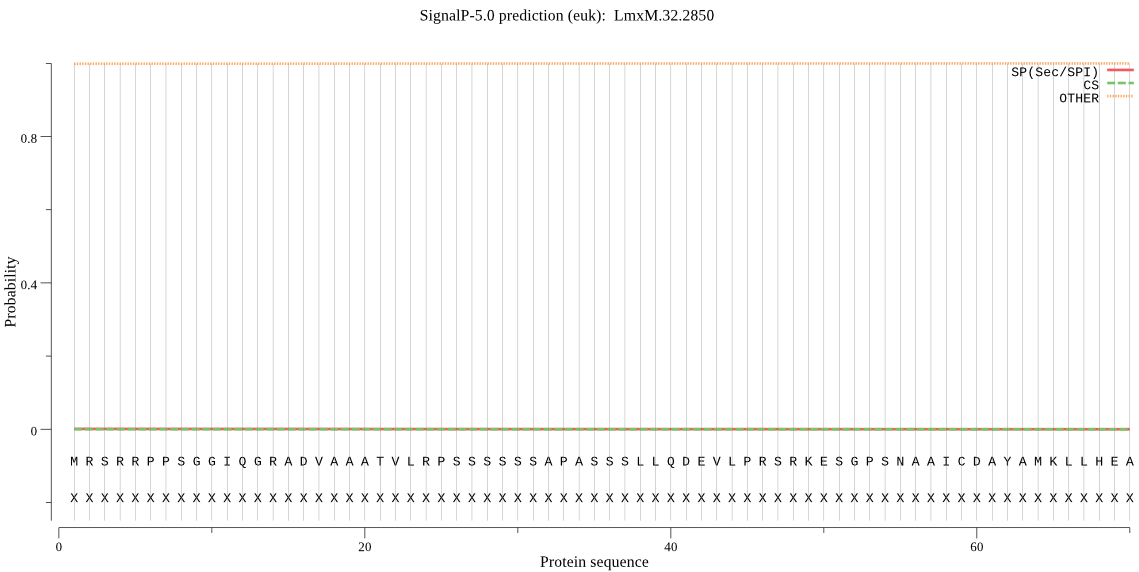
| LmxM.32.2850 | OTHER | 0.996650 | 0.002153 | 0.001197 |
MRSRRPPSGGIQGRADVAAATVLRPSSSSSSAPASSSLLQDEVLPRSRKESGPSNAAICD AYAMKLLHEATEVERCRGPHWRARCSPSILVALLIWATIGLVVYMKLDSSHLRPGAAPAT TSVETPLSQLPESCRGHHCLQKGGREPFGAVLGAHDGVFAYSNCNSDTCISSIQYQMAIP LPPGARTALDAPHATTRFMKTGMKWQCVEYARRYWMLRGKPAPAFFGAVNGAADIWDSLT HVTFLDNATTAPLLKFKNGASLGYGGGAPHVGDLLIYPRDSAGIFPYGHVSVVVEVEMTT KVKADDSYMAATAASPKPRQRHGLVYVAEQNWDSVTWPDPYHNYSRSLPLVVLESAEGRP LQYTIEDSFHGIQGWVRYDGEAYG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmxM.32.2850 | 51 S | SRKESGPSN | 0.994 | unsp | LmxM.32.2850 | 51 S | SRKESGPSN | 0.994 | unsp | LmxM.32.2850 | 51 S | SRKESGPSN | 0.994 | unsp | LmxM.32.2850 | 26 S | VLRPSSSSS | 0.995 | unsp | LmxM.32.2850 | 47 S | VLPRSRKES | 0.997 | unsp |