

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.34.4020 | OTHER | 0.994507 | 0.004982 | 0.000512 |
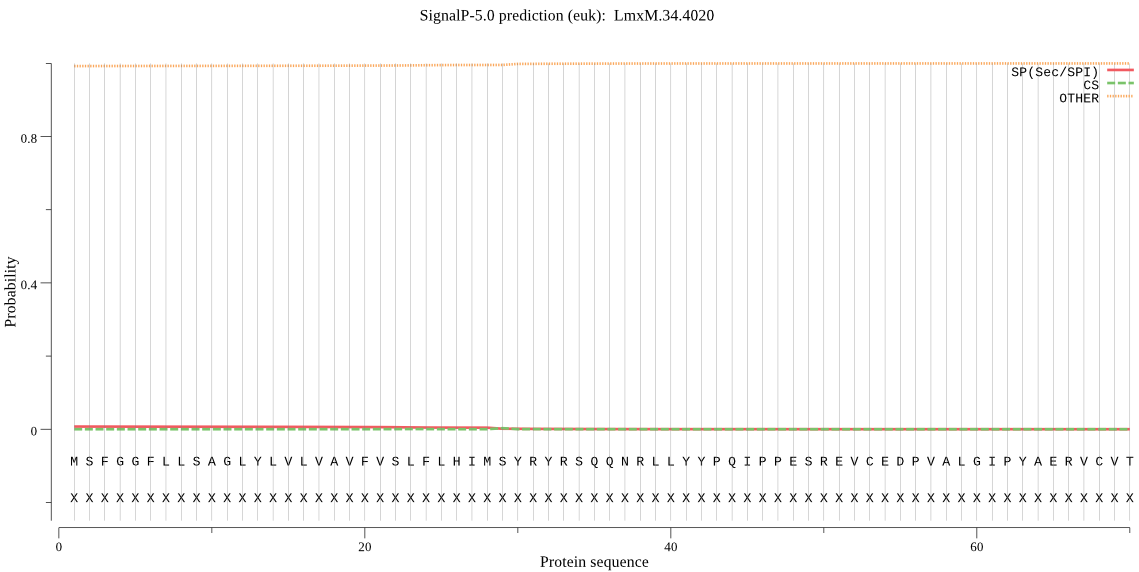
MSFGGFLLSAGLYLVLVAVFVSLFLHIMSYRYRSQQNRLLYYPQIPPESREVCEDPVALG IPYAERVCVTTADKVRLWGYMLWPAPAPSAEKSGNASVSDSIGHASSNLATAEGGVHVEV DASGGATESTSAGSTLPGSSRSVTMPSGMPTFVMLYFHGNAGNVGHRLPLARAFVTHLKC AVMMVDYRGFGLSDDSEQTQETLELDAQACFDYLWQDPRVPRDRIIVMGTSLGGAVSIHL AANERYARRIAAVIVENSFSSISDMASALSRPILTKLASQCPGLAVGIFEYYVKPLALRI SWNSAQKITKVVVPMLFLSGMRDEIVPPEQMRTLYKAATKCLRDGNGSDLTVPLRRFLEF EDGRHNNLPLMPGYMSALQDFVTDVRNAGSAAVV
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmxM.34.4020 | 97 S | SGNASVSDS | 0.996 | unsp |