

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.34.4850 | OTHER | 0.999837 | 0.000095 | 0.000068 |
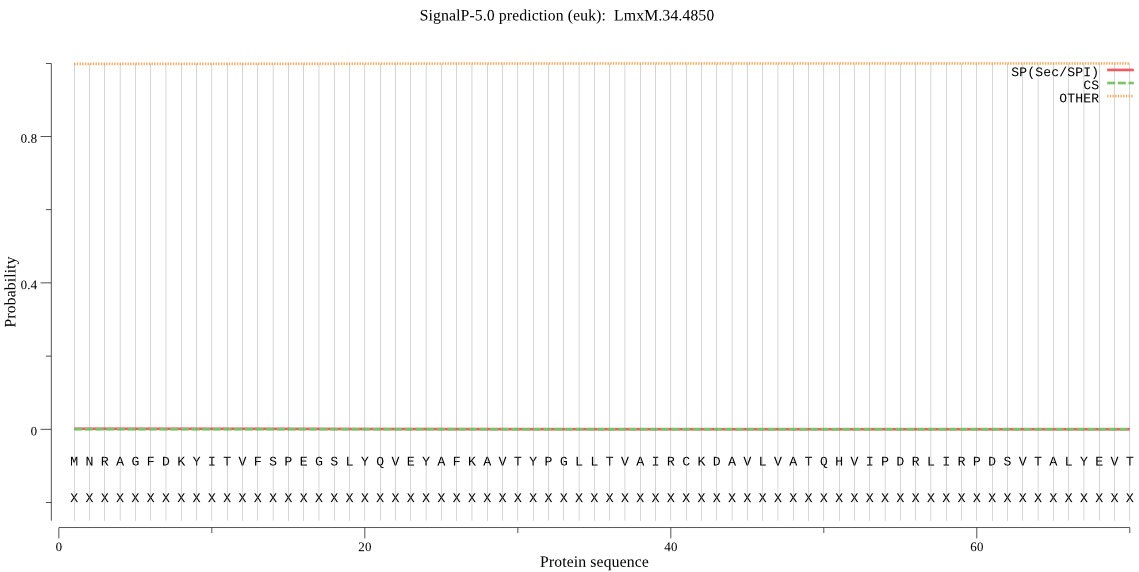
MNRAGFDKYITVFSPEGSLYQVEYAFKAVTYPGLLTVAIRCKDAVLVATQHVIPDRLIRP DSVTALYEVTPSIGCCMTGRAPDGRALVQRAREEASDYHYRYGMQIPIAVLAKRMGDKAQ VRTQQAGLRPMGVVTTFIGMDQSDQDGSLKPKIYTVDPAGWTGGHIACAVGKKQVEAMAF LEKRQKNTDFDDLTEKEAAMIALAALQSAIGTAVKAKEVEVGRCTAANPTFQRVPNSEVE EWLTAVAEAD