

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmxM.36.0200 | SP | 0.378462 | 0.610118 | 0.011421 | CS pos: 35-36. VSC-AV. Pr: 0.5455 |
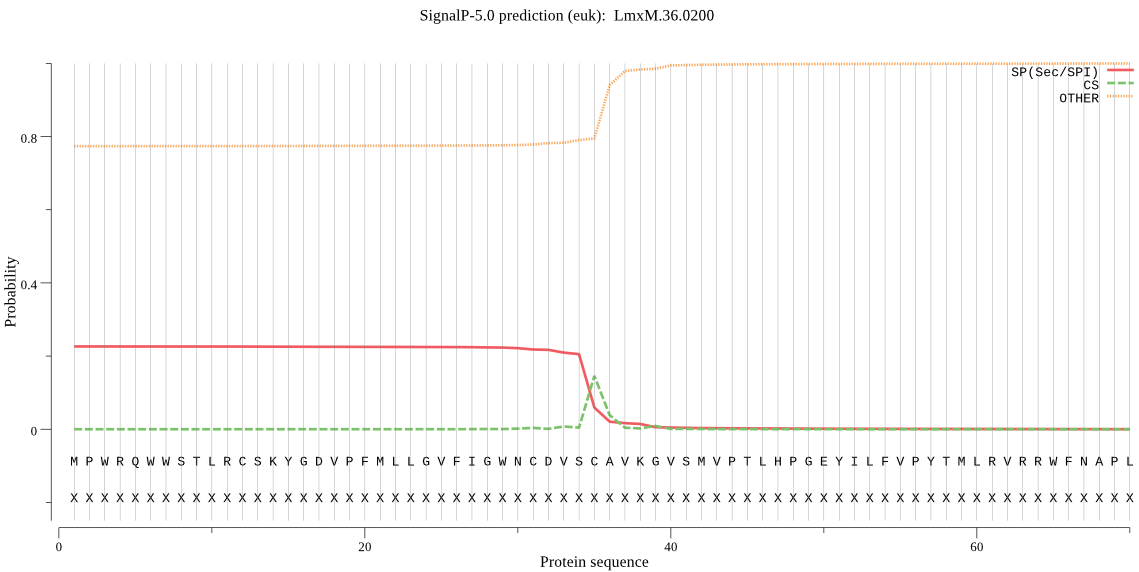
MPWRQWWSTLRCSKYGDVPFMLLGVFIGWNCDVSCAVKGVSMVPTLHPGEYILFVPYTML RVRRWFNAPLVNLSDVVVVKVSDDLSVCKRVVKCTSSRAQAEEWGKDHYVEVVPAPYSSP AAQHINGHAEDSTEMDSVTNSERAYFDYVARNTVRSKDWDSCTDRIPNPSQWVWLEGDNK TDSFDSRRCGPVPVECVRGLVLASIWPSPHTLQRPPPPPRASQLP
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmxM.36.0200 | 186 S | DSFDSRRCG | 0.994 | unsp |