

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
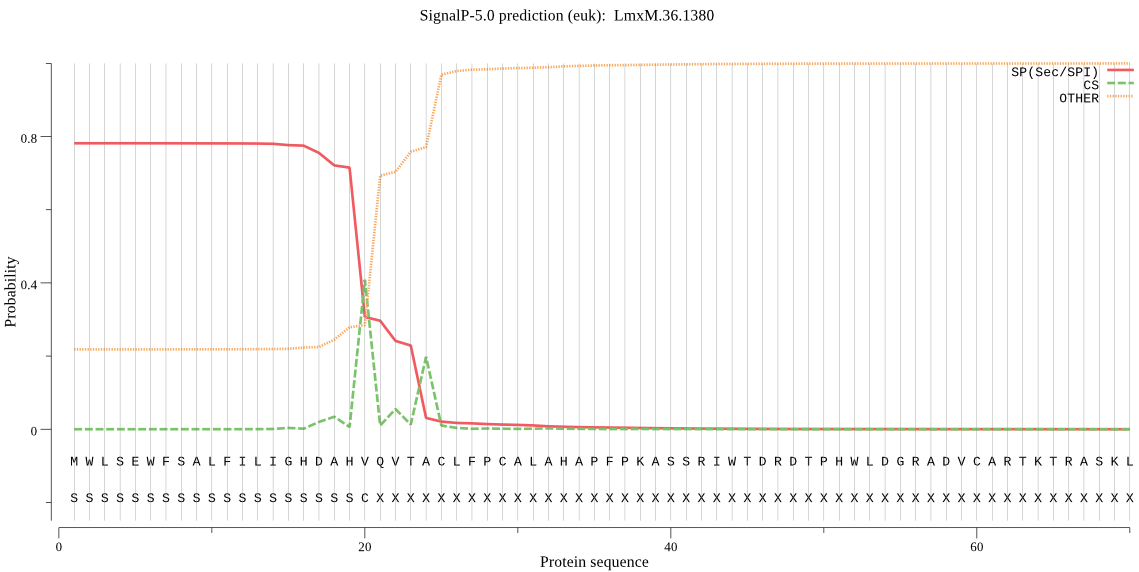
| LmxM.36.1380 | SP | 0.137315 | 0.860526 | 0.002160 | CS pos: 24-25. VTA-CL. Pr: 0.4715 |
MWLSEWFSALFILIGHDAHVQVTACLFPCALAHAPFPKASSRIWTDRDTPHWLDGRADVC ARTKTRASKLVPMTSTLTNSNRINIPAVCRLRERNTSKTVGSGVLVAPGLLLTSATAIPT QSRATHVVATFFEGTKKNAVDVPLQPQRYYFASRYPPEMDYCLISCEEAPLFNVTPVHMA LTAKGWAPVEEGDVVLIVEHPIGGVFGTAAAPLNGKATSPSTLSETGESNFDSVQAPYVE QKRFEEVLRCRGNVSFLKSNGSWRTAGCPAFNEHGHLVGLQSQSRADGEGVVNRILSLIS IVKHLFANVQLPHLPQEDITFEDVWDTWYVTNDITRIIAILANFKGKSMAQQVTWRLCEL TAKPQLVKSIASNGGIEVILQNLSLFCSDEKLSEIGLRALWNVSIGEEAHLAAIIKREGV HTILNVVESFPANETVLEFGAVLLHNIAGCRAAPNFSEAHGARALKALYPGFQQFRESVV LQKFALSFFATLARLNEEFACNLVQQDVIDHIIHLIEEKPKQIFLMEIVVGFVAELAQYP QTVELMCGKAPQYAAASMTYLIDLLIHIILEYKDLDSLLVSGNRALWGLGIVPTCRGIIL ENPRGIDALRISLPALIASTHR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmxM.36.1380 | 97 S | ERNTSKTVG | 0.993 | unsp |