

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
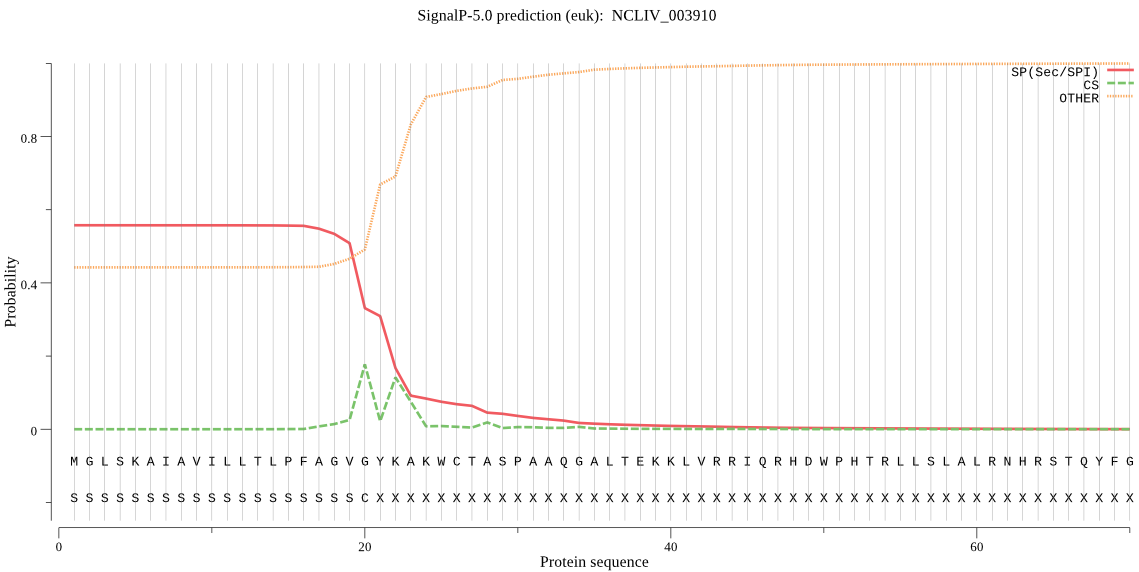
| NCLIV_003910 | SP | 0.030431 | 0.964861 | 0.004707 | CS pos: 20-21. GVG-YK. Pr: 0.3780 |
MGLSKAIAVILLTLPFAGVGYKAKWCTASPAAQGALTEKKLVRRIQRHDWPHTRLLSLAL RNHRSTQYFGEISVGTPPSLFKVVFDSGSHQFWIPSKECQSSSCRTHSRLDCSRSSSCRD HGNTGWNESVAVTFGTGRIVYRKALETVKIGDVVIPSQAIGLVVDQTDEPFANLPFDGIV GLGECSLLYTRVPLLSGSRLAGSTELTGEKDLLDNMKNEGIITDRVFAFYLSRRSALGGV ISFGGFDPRFVQPGKPIQWIGMLPQKGWAMPLIDFKVDGVRLDLCFDSAASRCAAVLDTG TSSIGGPRADIHHILTMLGAAPRCERKLAMKSLTVIIEQEAGREIEFELTPDDYVVDNLK PSDDSTSCPAAFMPLELKRHPVRTFVLGEVFIRKFYAIFDRENKKIGECTKHRRHAYTWA LSFPFLSRPCRSCYRREA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_003910 | 116 S | CSRSSSCRD | 0.993 | unsp | NCLIV_003910 | 117 S | SRSSSCRDH | 0.997 | unsp |