

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
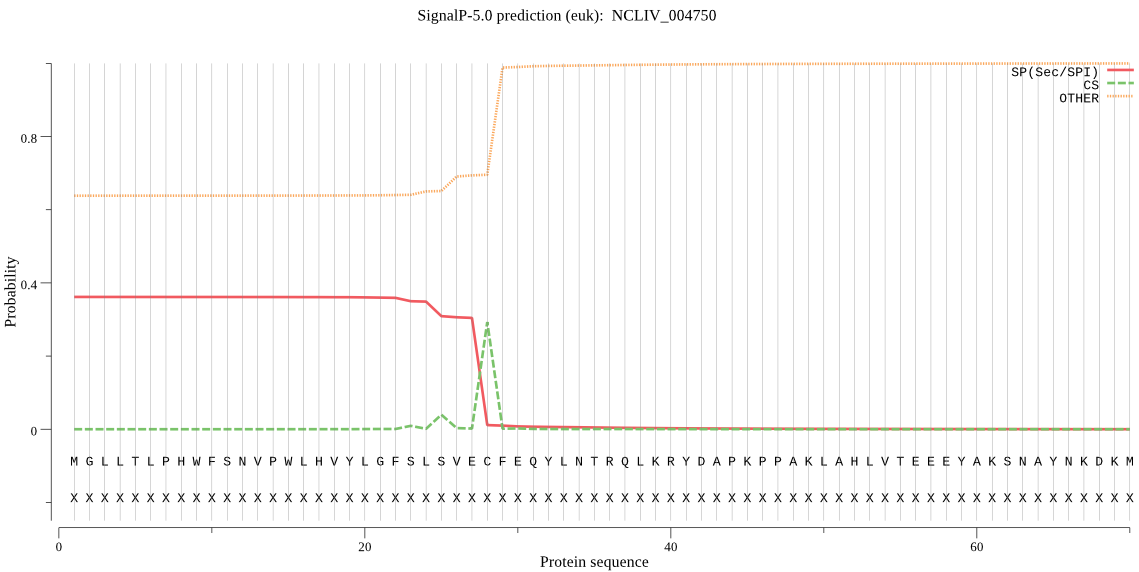
| NCLIV_004750 | OTHER | 0.594581 | 0.401178 | 0.004241 |
MGLLTLPHWFSNVPWLHVYLGFSLSVECFEQYLNTRQLKRYDAPKPPAKLAHLVTEEEYA KSNAYNKDKMRFGIFSSLFQTSISLISTACFLGPYLWRLAGTLVGKNGNEYTQSLVDLAL SAVIGECISTPFQLYGDFVVEEKHGFNKKTLALFFKDKLLSLGLTSLIGGPVAYAAIWLI NVSVLTQLWGFSVATVIAMMFIYPNLIAPLFNKFEPLKDEELRGKICDLAKKLDFPLTKL YEMDNSKRSGHSNAYFYGFWWSKRIVLYDTLLHLPHDQILAILGHEMGHWKKNHTTKMMA VNFLQLFCTLYLFGLVMGSDALFDSFGYTDTRASVVGLKLFSNIFLPVNTLISLMMTIYS RKNEFEADAFACELGYSEPLKQGLVAIHAENKSCLDPDPWFSFWHYSHPPLLERLRAIEA IQEKEEKEN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_004750 | 334 S | DTRASVVGL | 0.995 | unsp |