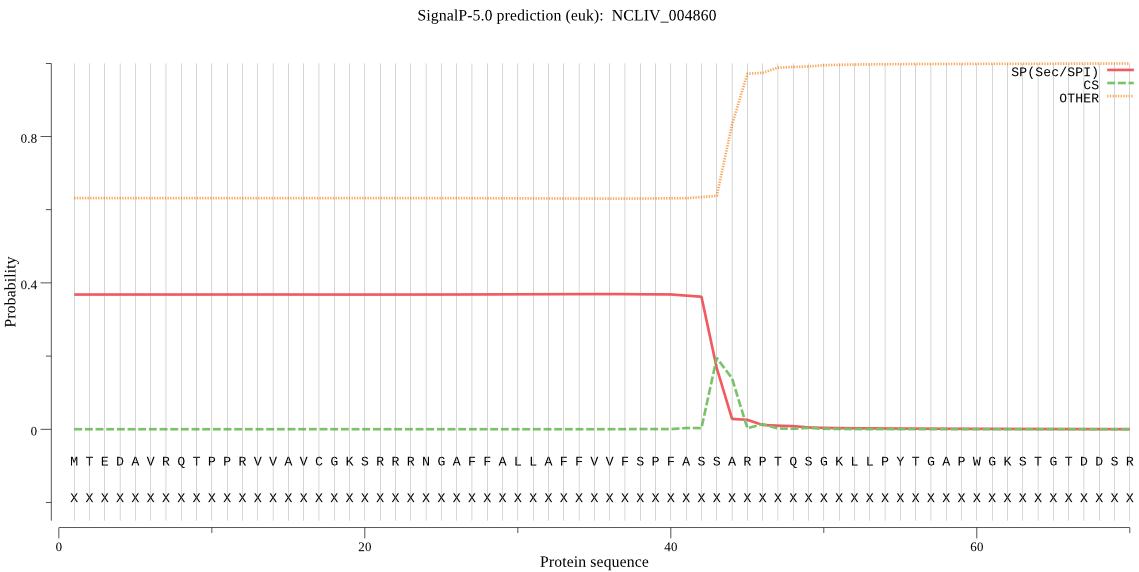
Fasta :-
MTEDAVRQTPPRVVAVCGKSRRRNGAFFALLAFFVVFSPFASSARPTQSGKLLPYTGAPW
GKSTGTDDSRRVFVSPASLRPIFSAPLASPGPSHVGQPKGIEFGASPAAPRNPSPAISAV
SAHGLRRRRPLLPFSHFRRSLAFLSGPNLSASSSTSSCSPSRSRRAPAGPPEALDSFRCH
RTSLSGVESPELRQSTFLRPPSARSSLSPASLSSFSVSSSADHAGGLQTALHAATSEARV
ENRERVSLPRVDGDGTEGDILANYVRKHGGKRVIRRVLLANNGMAATKSIFSMRQWAYME
LGDEKLLEFVVMATPEDMRANPEYIRRADKIVEVPGGPNRNNYANVDLICQIAAQEKVDA
VWPGWGHASENPNLPRRLRELGITFIGPSATVMAALGDKIAANILAQTAGVPSIPWSGDS
LQATLDDTGAIPREVFERATVQSAADCEKVAARIGYPIMIKASEGGGGKGIRMVDRKEQL
RGAYEQVVAEVPGSPVFMMQLCTAARHIEVQILGDEDGQAVALSGRDCSTQRRFQKIFEE
APPTTVVPPHTMKEMERAAQRLTQSLGYVGAGTVEYLYNRKDNKFFFLELNPRLQVEHPV
SEGITGVNLPAAQLQVAMGIPLWRIPDIRRFFGRDPNAGDRIDFMKDDYLPIERHVIASR
ITAENPDEGFKPTSGRVDRLEFQPLENVWGYFSVGASGGVHEYADSQFGHIFATGKNREE
ARKKLVLGLKRVDVRGEIRTPIEYLVELLENEDFIENRIDTSWLDKLIKQRKSLGEKKVR
KAHVVLAAVLFRAIGALKEKEQRVLSALQRGQRYIHDLAALNTVSQEIVYEDQRFAFQVS
RTGPQLYLLKLNAQEVECLVREQPDGSYIVLFPGEKSHKFNGREEPLGLRLQVDGSTVLL
PNVFDPSELRSDVTGKVVRYLVPDKSQVNKGEPYVEVEAMKMVMTLTAGETGVLSHTKSP
GSVINTGDILGTLELEDPSRVKKIVDFAGVLDLPGAKKRAGEAEGEAEREKEKGRQALSL
FGSPEEEAAMRLRLAMEGYEQDVEPTVQKLAFPSGNAENLKVKKEKLTELVVELFQSFLG
VDEYFASLQGSEAGVEGPAEGGKAQDPATLLKMKLAHSHLQPRIQLVLSLLRALQNFPNF
FPDWTMPVSLELCLKRLSALHGRAYGQIALEAMRLLRSFRVAPFDQRVQRLKEQLLQIPA
LSASASEAERREAVRQAERIARSSTTSAGIDLLSHLFGDEQVGLRALEVAVRRHYRGFPI
YALHTELFEVKTRDERGEKTRKVPVALWSFRQGDLTGDEAPLRTGVMGAFESLQEVQEVG
DRLLARLQQFDAEKKGTGTKVRREAEGEKGETDAETEPRNVCLFALTTPINGVGAANAET
EDRSSPSGEEKLKTQMEGILRTLQPSLRSADVRLAAVHVPQVKKSCRMFNFLASDGFNEC
RIRRDLCPTVPSLIELKSLQTSFDLTRLDALDPNTQAYLALPTVAPPAQTGASPASPAPA
AVAAAPAKKRLSLAQAPQSVFVRRLIFENEVKSDLSDLPRILLDLLDVLDRCANDPRVSA
TASGRLFLHIVSPLNLTSAKHVTEYFQKVMKRFRSEHNERLLRLRVDQLEVKMHLRKGQG
DKGRGSDREGPDADAHEREKEGLQVLRLSVSSHQGSWLHTKVAEDVPHVLTGDPIAHRRF
VFSEDGEREAAAGPAEKTTHKSLESGPATGAGGTAEAEGREGKATPGSREASEAEDLPTR
VSVGLKTLRGIETGASAEGGERPVASPSLSSVNWEEGDRKKQQVLSLSPGQKNEFFQHER
NVEPYPEVDRIAMARSAARRAGSTYIFDFLGLLEIALLQSWEAHLKEKGGKEGGAGGWDE
AVPRDLFKAEAFRLSPKGDLYLDPDWRVADNKIGMVGFLVTLKTPEYPAGRQLVLLGNDI
TFQGGSFGVPEHRFFTRVSRFSREQGLPRVYIACNSGARIGLYENLKDKIKVEWKDASNP
SLGFKYLYLSAEDYEALPSGVVSGHFEDNGKSDSGDGRRFVLDAIIGDADKFIGVENLRG
SGTIAGETSRAYDETFTLSYVTGRSVGIGAYIVRLAQRTIQMVRGPLLLTGYQALNKLLG
REVYASQDQLGGPEVMFRNGVSHLVVQNDQEGMKEVLRWLSYTPKTARDSVNSANLFSVD
PVERDVDFTPTRAPYDVRHMLAGYTKEDGTFVTGFFDKNSFKEYLAGWGKSVVVGRARLG
GIPFGAIAVETRTTEARVPADPSSPDSRESVIMHAGQVWFPDSAYKTAQAINDFNRGENL
PLIIFANWRGFSGGTRDMFEEILKFGSQIVDALRTYKQPVFIYIPPHGELRGGSWVVVDP
TINLQKMEMYADENARGGVLEPPGICEIKYRAADQKALMHRVDDVLKELDRQLQDCQTAS
DAIDLKEKIKRRETALEPLYLSIARFYADLHDRPERMKARGVISSVVNWKKSRAFFYWRA
KRRLLQDDLEARILAADPRLDFMEARAKLEGLLASRGVDISSDKAACEFLDSAQGKEVTQ
AIVEQSRREGAIEKIRYILSSLPESARRVTLESAASPTS