

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
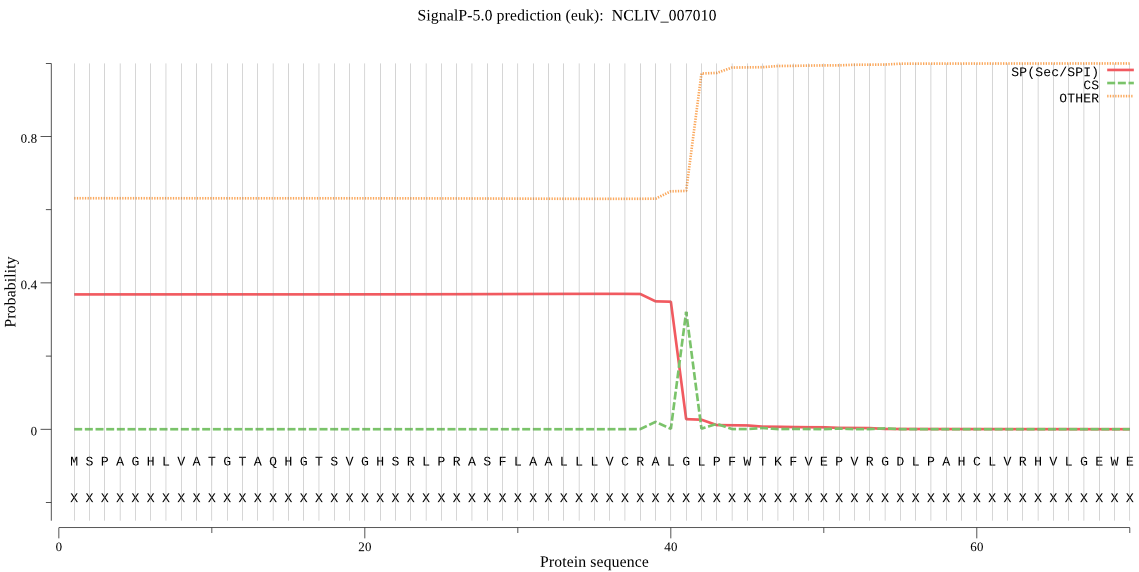
| NCLIV_007010 | SP | 0.345013 | 0.651638 | 0.003349 | CS pos: 41-42. ALG-LP. Pr: 0.9100 |
MSPAGHLVATGTAQHGTSVGHSRLPRASFLAALLLVCRALGLPFWTKFVEPVRGDLPAHC LVRHVLGEWEIQEGLWLPCKGELTAAAAQLHDPYCGYGVPDKIDAHDPMTPPNISPSFHL LRTSRFTLHPDFRVDFDDGSTGVWTLVYDEGLHFEVTTPQSRRFFAFFKYEVAPDGRNLA WSYCHTTLVGWWDRVPQGDAVRQSLEPTPPAYDVPELVVPETPADQLLHSLKRGCWWGRK VGEDLSVPTNEVPKERRTPLDVPIWPTSLPPDIQSVAAVVRKSLSTDAPWEPVEDDYIEI RGRKLRSIQEVWENAGNQPLLMSRQHIEGRRATVKASLPLHVLSGVQPFGSPAGSEPPWR TIREFDWSNEEHVALRIGRRRSVVPDAPNQGRCGSCYAISTGTVLTSRLWIRYAANDDVF GKINVSAFQGTSCNVYNQGCGGGYVFLALKFGQEHGFRTEECVQEYTREAHHRSSPLSPS LQTCHDLGGQLGTSAYGCRAPPARSSLPESCSVSIKVASWQYVGGVYGGCSEDAMLRTLW EHGPMAASIEPTVAFTVYRKGVFKSAYNSLVQKGENWVWEKVDHAVVVVGWGWARHGDKW LPYWKVRNSWGARWGEGGYARVIRGVNEMAIERVAVVGEVSLFRDGKHIPPTPAKEPTTS QLAANSSTEPSGNREFSVETLPTLRGVTVMRAARTHQPSEDDREAGESESEAASEQAGAP EDADESRIDAKFAAPQPSFLSQSVKRHRT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_007010 | 368 S | EFDWSNEEH | 0.994 | unsp | NCLIV_007010 | 368 S | EFDWSNEEH | 0.994 | unsp | NCLIV_007010 | 368 S | EFDWSNEEH | 0.994 | unsp | NCLIV_007010 | 382 S | GRRRSVVPD | 0.996 | unsp | NCLIV_007010 | 475 S | HHRSSPLSP | 0.993 | unsp | NCLIV_007010 | 478 S | SSPLSPSLQ | 0.995 | unsp | NCLIV_007010 | 506 S | PARSSLPES | 0.992 | unsp | NCLIV_007010 | 677 S | NREFSVETL | 0.994 | unsp | NCLIV_007010 | 699 S | THQPSEDDR | 0.996 | unsp | NCLIV_007010 | 708 S | EAGESESEA | 0.995 | unsp | NCLIV_007010 | 726 S | DADESRIDA | 0.996 | unsp | NCLIV_007010 | 743 S | FLSQSVKRH | 0.992 | unsp | NCLIV_007010 | 204 S | AVRQSLEPT | 0.995 | unsp | NCLIV_007010 | 230 S | QLLHSLKRG | 0.995 | unsp |