

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
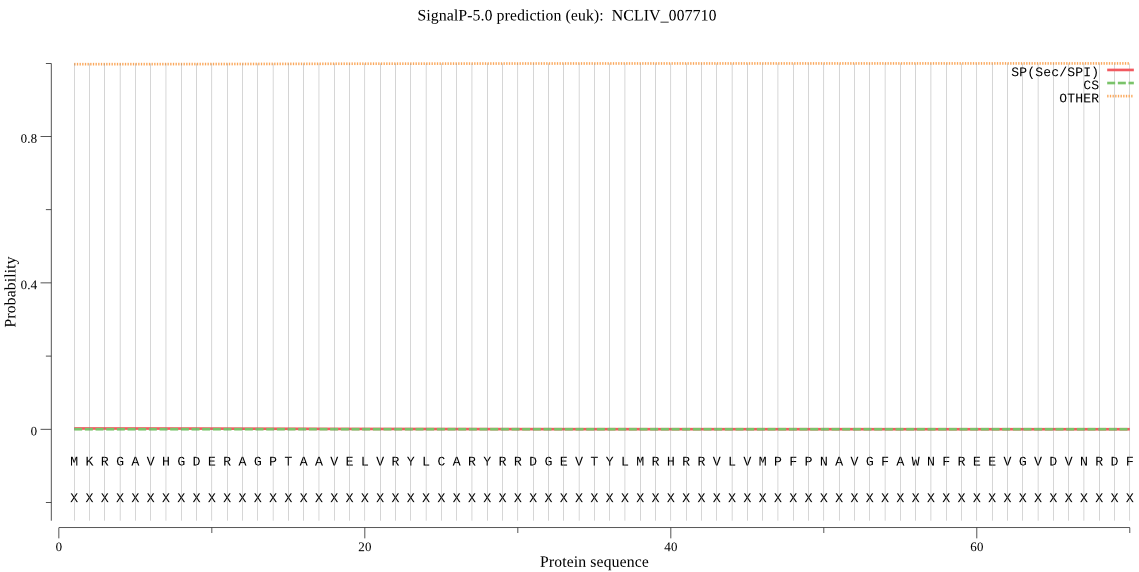
| NCLIV_007710 | OTHER | 0.999582 | 0.000188 | 0.000229 |
MKRGAVHGDERAGPTAAVELVRYLCARYRRDGEVTYLMRHRRVLVMPFPNAVGFAWNFRE EVGVDVNRDFPYQRQADQCFQSVAARAISELFRTHLILGGITWHGGMRAVAYPWGSYDHS QQLGREQWQSRPSPDDEAFKSLAGALQRAGGRDLEKGDFYYPVGSMTDLVYPVNGGMEDW AYGASFEPSPDPITVCEPAPYELAVASSLSPSAPSTASASPAALASRLPMLKQSEARAEG DRFRSDEETTQIQADDSANGGDRPGAEGGARRLSGRSFAHAGEKGSNRNRGTTTMPLRGE ADDGVPEGMALGGEASEEGGETESAQNGAEGKREGEAERKAKSGKGRDDADPEAKWKTAD RDGVQLYAYPRSRSVYSAADIRCALFLVEMHDDKGPSRVSMGPRPVDRPTLLLPDEGGDG VQKSAVARASEESDFLTIRNVRMALKFIEKAQPDLLFTSTPPLYQPPGAVSIFAFYPVGC NVLSHTELQIRRGRCEDLLSVSHSSRLGPELLPAWHAAEVLVREVGKDLPTSCRELGVWE RQGSLAEDRDRPPERFEWWGGGGNKDIALNQGSVEQKRNRVEIQYKIPPEALDGDYCSVV FASFDQDYEARSSEGNAVIVGHKTWLFPAAIPALFPVVSAAGYPLIIRGDNVEGFARLDF PLEAFRLSSKTSEAALKRDTTSDTTVQLFLNFGGNFNEERFRYDPFTGVLRYPKATNPSL SIRRVDLSVSLSRPFSSLPPGRFLFTVFEDGDLLQHTVNQFLFPASSLSSLSASVSSFSV SSSGSPCSAKSRATQGNFYSAMLLGELPTGTMGPSELERLQKTRMANFVKRQKERTESTW GGRPHLSSTWSRQVAEKLKQAIAPAVVDFSLIDAKTLRGQADVPVTALLGRTVVVQWIPE SSRGSDKKEEKEGEQRGAQEGDAARADTGLSKKTGTSRVASSGYRPSQSVMAVVGASVHT PPIIHSIHEKLAPLPADPTQRSASSSPPFVSSLACFFRHEPARGERAGASLGREKPELTG ARHLAGGRGSREREEGPDTAGGSDRKVPEQQMGEEGGRRAEETASTEKREAGDMEAAPEA KTTVWEGERERDVEASERDAARTLLGNLIGYLRLTYTVDSVGEQRKENLEEGGGSGAGRV RVEAELCETSMGIPADAPLYLKFHEDCCPPLTMYPSTRQRLQFASQAQRVPEQRQSWLGR FLGVRAGDTANTKTAMPPEDETGSWELPEGTANLLSLRNCRCGPGTIVTLATEVFASSLH ASSSFSPFSSNTFFLPPADMVDSAHRRTLACALGLVPPVPGQSSEASRHDETSNRLPFSV SLTGGEPSIAASPFISRFICGSSSRPSWRQWSKRPEAAARRSASTESNKEGHQTQAEEDV ADFIAFFEAAEQAREDHDMPPKYHLVIASLIALIGLVCILSPCLISVYDCCLSHHGVGGM QLSERPSERRSPFNEYPEIHAAEGEIPEIPEGEEESPGGSASHTVDRFGRLVDAFAEANS ELNGDRQAKKEDGKAIVAKSNLFSSRLRGVTDDEVFNVPSSLGHAVVSTAPRRLSGGTGK RASYEPFFSIAGEDDDSEEDEGGFQH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_007710 | 274 S | ARRLSGRSF | 0.997 | unsp | NCLIV_007710 | 274 S | ARRLSGRSF | 0.997 | unsp | NCLIV_007710 | 274 S | ARRLSGRSF | 0.997 | unsp | NCLIV_007710 | 343 S | RKAKSGKGR | 0.996 | unsp | NCLIV_007710 | 430 S | VARASEESD | 0.997 | unsp | NCLIV_007710 | 532 S | DLPTSCREL | 0.993 | unsp | NCLIV_007710 | 612 S | YEARSSEGN | 0.992 | unsp | NCLIV_007710 | 613 S | EARSSEGNA | 0.994 | unsp | NCLIV_007710 | 668 S | AFRLSSKTS | 0.995 | unsp | NCLIV_007710 | 721 S | NPSLSIRRV | 0.994 | unsp | NCLIV_007710 | 870 S | VVDFSLIDA | 0.993 | unsp | NCLIV_007710 | 905 S | SSRGSDKKE | 0.998 | unsp | NCLIV_007710 | 984 S | QRSASSSPP | 0.996 | unsp | NCLIV_007710 | 1030 S | GGRGSRERE | 0.997 | unsp | NCLIV_007710 | 1096 S | DVEASERDA | 0.997 | unsp | NCLIV_007710 | 1120 S | YTVDSVGEQ | 0.997 | unsp | NCLIV_007710 | 1307 S | SSEASRHDE | 0.992 | unsp | NCLIV_007710 | 1347 S | SSRPSWRQW | 0.997 | unsp | NCLIV_007710 | 1364 S | RRSASTESN | 0.997 | unsp | NCLIV_007710 | 1367 S | ASTESNKEG | 0.995 | unsp | NCLIV_007710 | 1447 S | SERPSERRS | 0.997 | unsp | NCLIV_007710 | 1451 S | SERRSPFNE | 0.997 | unsp | NCLIV_007710 | 1555 S | PRRLSGGTG | 0.996 | unsp | NCLIV_007710 | 1563 S | GKRASYEPF | 0.997 | unsp | NCLIV_007710 | 1577 S | EDDDSEEDE | 0.998 | unsp | NCLIV_007710 | 133 S | QSRPSPDDE | 0.998 | unsp | NCLIV_007710 | 245 S | DRFRSDEET | 0.998 | unsp |