

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
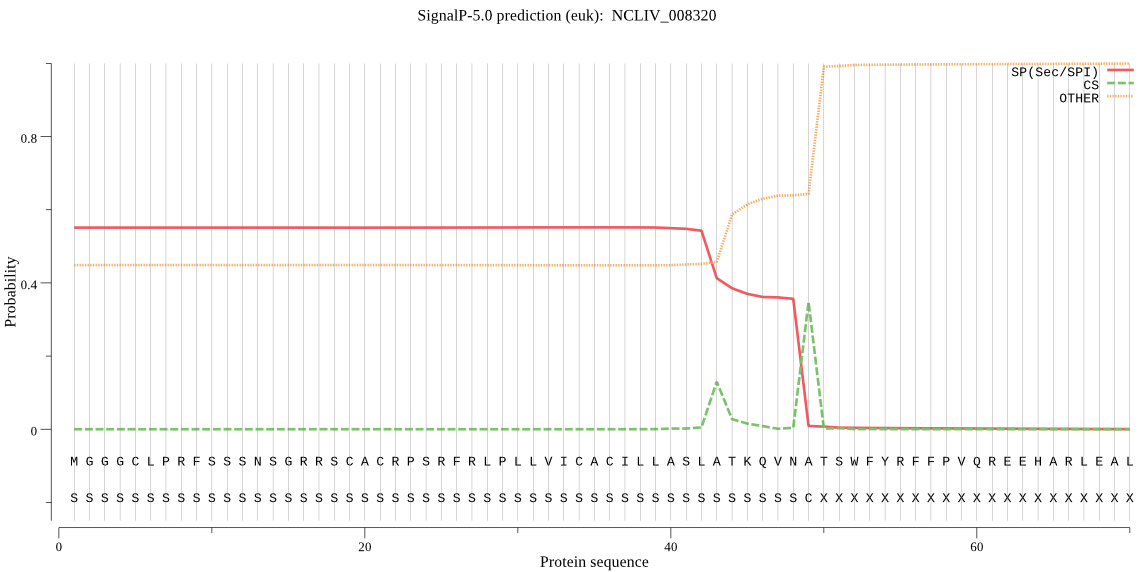
| NCLIV_008320 | SP | 0.272023 | 0.724540 | 0.003438 | CS pos: 49-50. VNA-TS. Pr: 0.7012 |
MGGGCLPRFSSSNSGRRSCACRPSRFRLPLLVICACILLASLATKQVNATSWFYRFFPVQ REEHARLEALKQGRHTGKEEIDETEGGREIREFEEGTGSRDASAPSAEEGILPAEFLPPF LRRFLPPRTSQAAQVNVSRENERRSTVREDIEVKEGWFEQPLDHGNPLVFNRPQHAAWRQ KFYSAKRKRPSCQKREQARNEHGSLENAERSEQGQACPDDAIRPVFVYIGGEGPLSSMEV KQGLLAEMGEAFGASVYALEHRYYGDSHPRPDSSVPNLQWLTSHQALGDLAAFVAHVKRE QAEQHPQNVSPEDIPVIVFGCSYPGSLAAYARSKYPASILGAISSSSPVEASALFQAFDQ TVQRVLPSACTSQIKAATAIVERRIFTSEEEAVKVAAKFGCGAEVSMKTHDQRVALLYVI ADAVAQSVQYNRNPKRPWIDEVCNCFAEAPPARTEKVGNEDTKARRSEEDELLDALAKAV QLMLAELKMTCKDSNMLQLADTRLGPQASASARLWVWQSCAEYGYWQVAYKGSVRSRLID LNWHLRMCDALFPLPSGSKFSTDVVDETNVWSGDKHVVGVGAATNIHFTNGENDPWAPLS VTEISPFVTERQGLSSFTIKNGSHCNDFYAYEDGTEPLPVTEAKARIQRAIRLWLEDFQA RHAQRRARNTPETNKGISRGENEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_008320 | 138 S | QVNVSRENE | 0.993 | unsp | NCLIV_008320 | 138 S | QVNVSRENE | 0.993 | unsp | NCLIV_008320 | 138 S | QVNVSRENE | 0.993 | unsp | NCLIV_008320 | 145 S | NERRSTVRE | 0.997 | unsp | NCLIV_008320 | 146 T | ERRSTVRED | 0.997 | unsp | NCLIV_008320 | 184 S | QKFYSAKRK | 0.993 | unsp | NCLIV_008320 | 204 S | NEHGSLENA | 0.992 | unsp | NCLIV_008320 | 310 S | PQNVSPEDI | 0.995 | unsp | NCLIV_008320 | 347 S | ISSSSPVEA | 0.996 | unsp | NCLIV_008320 | 388 S | RIFTSEEEA | 0.996 | unsp | NCLIV_008320 | 467 S | KARRSEEDE | 0.998 | unsp | NCLIV_008320 | 678 S | NKGISRGEN | 0.997 | unsp | NCLIV_008320 | 14 S | SSSNSGRRS | 0.996 | unsp | NCLIV_008320 | 106 S | ASAPSAEEG | 0.996 | unsp |