

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
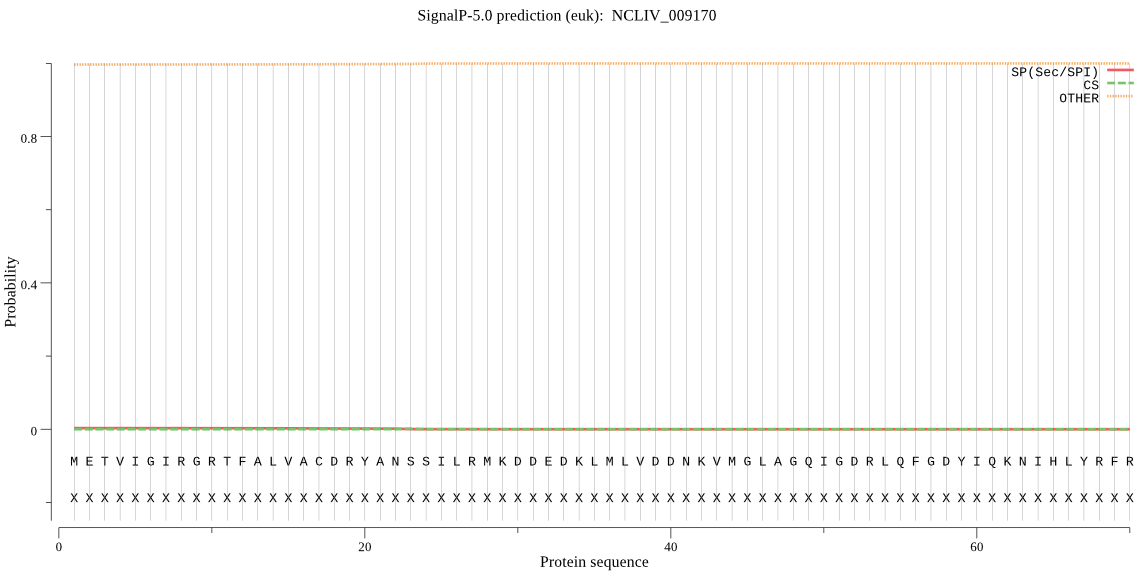
| NCLIV_009170 | OTHER | 0.986575 | 0.005678 | 0.007747 |
METVIGIRGRTFALVACDRYANSSILRMKDDEDKLMLVDDNKVMGLAGQIGDRLQFGDYI QKNIHLYRFRNNKKLSCAATASFTRQQLAYYLRRSPYHVDVMLAGYDEMGPHLYWMDYLA SMASVNKGAHGYAAYFLGGLLDRYYHPELTEEEALKIIEMCKKELMTRFVISQASFRVKI ATKDGIRTLDI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_009170 | 95 S | YLRRSPYHV | 0.993 | unsp |