

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_011200 | OTHER | 0.999999 | 0.000000 | 0.000001 |
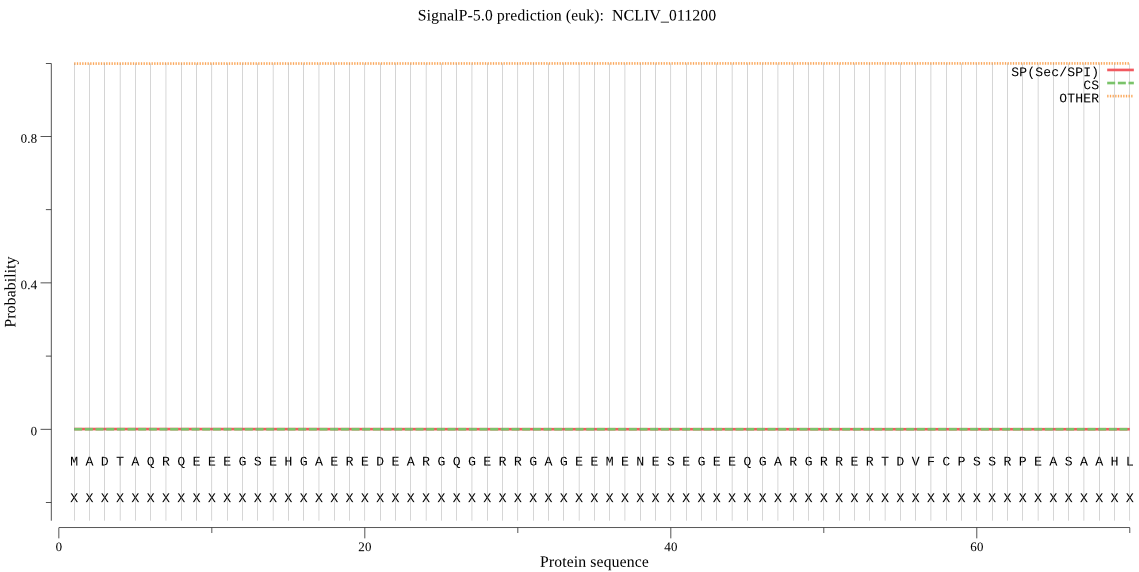
MADTAQRQEEEGSEHGAEREDEARGQGERRGAGEEMENESEGEEQGARGRRERTDVFCPS SRPEASAAHLRKRRNAKRTFLSSVFPDSLVSPSISSCEESLVPSRSSTRHCEEREKEKLL PVDILINSPGGSVTAALGILDLFQSLPFDVHTTCVGQAAGVAAMLLAAGTPGCRRAFPSS RLSLRQIEGRVEGQAEAIDRETLEIENVQRMVYRHLAAFCTGRQARRDEREQEREQERER QIEKREKAIREDCEKELFLSAGEAVKYGLIDRVIPVKYRPPWHTVGRPKAD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_011200 | 96 S | PSISSCEES | 0.99 | unsp | NCLIV_011200 | 183 S | SSRLSLRQI | 0.996 | unsp |