

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_013230 | OTHER | 0.999998 | 0.000002 | 0.000000 |
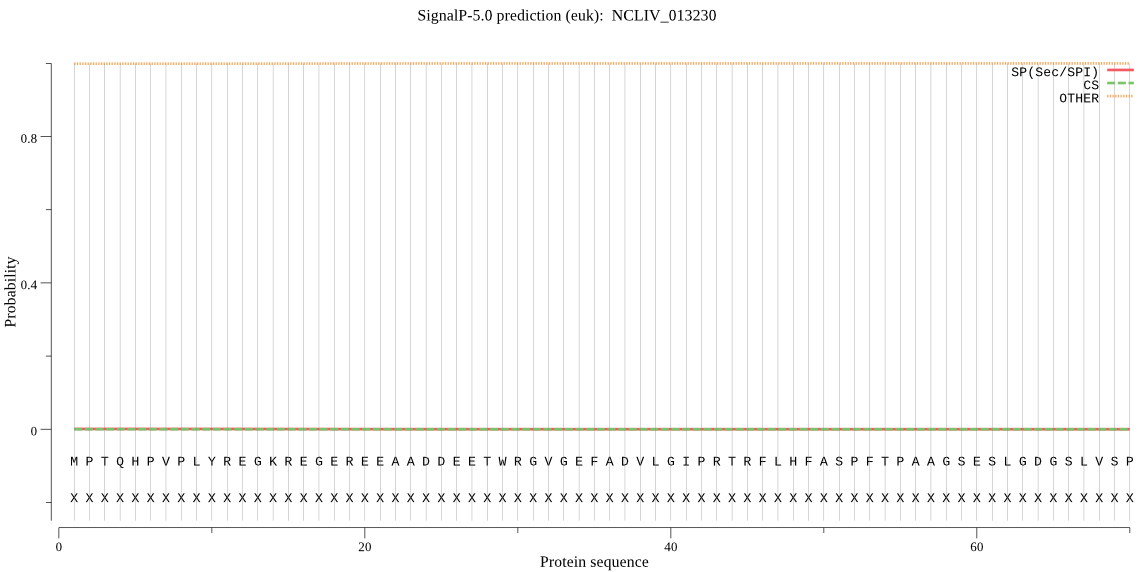
MPTQHPVPLYREGKREGEREEAADDEETWRGVGEFADVLGIPRTRFLHFASPFTPAAGSE SLGDGSLVSPFDTEWQPAVALLRGRALSVTPSPSCPDLSSPSSVLSSLPSSLPSSFSSSL PSSFSSSLPSSFPSGSAPSRASHAPSPPFCATVAVGLAWGTAALLLACGTRGFRAATRNA SIALRFPRVSLGQGKTAELRSRAEAVLSGQRAQAELLASLLGNAEKEKKREGGENEAQTE SRTSDSNATVSDLLRLMRKGAFLTPEEAQDLGIIDHIL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_013230 | 88 S | GRALSVTPS | 0.993 | unsp | NCLIV_013230 | 88 S | GRALSVTPS | 0.993 | unsp | NCLIV_013230 | 88 S | GRALSVTPS | 0.993 | unsp | NCLIV_013230 | 244 S | ESRTSDSNA | 0.998 | unsp | NCLIV_013230 | 61 S | AGSESLGDG | 0.991 | unsp | NCLIV_013230 | 69 S | GSLVSPFDT | 0.997 | unsp |