

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_014330 | OTHER | 0.999575 | 0.000359 | 0.000066 |
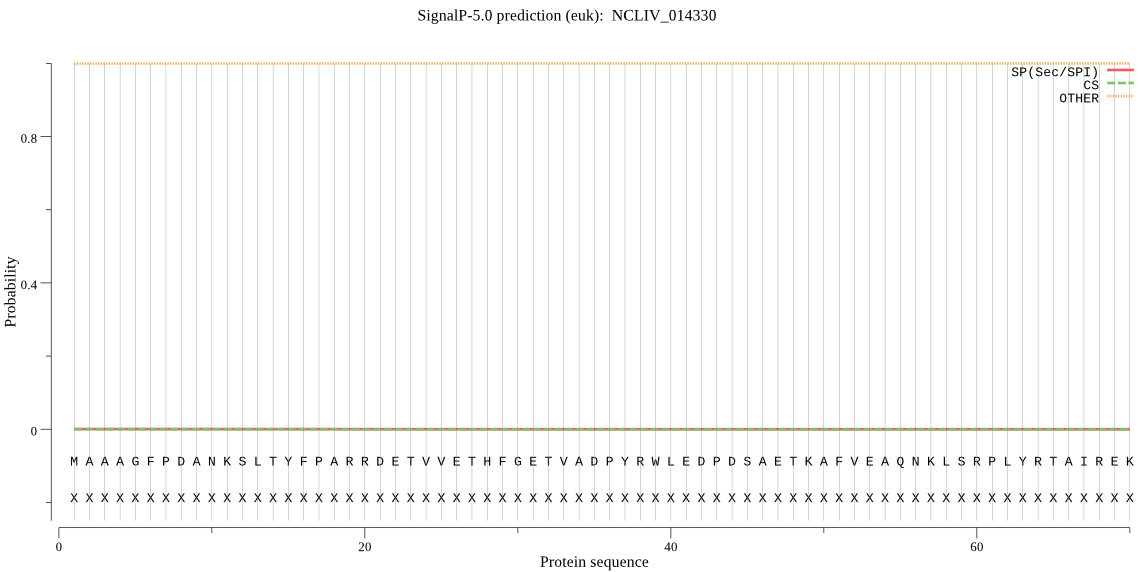
MAAAGFPDANKSLTYFPARRDETVVETHFGETVADPYRWLEDPDSAETKAFVEAQNKLSR PLYRTAIREKFFKKLEQVYDYPKYGVPFREGDMWYNWYNTGLQNQNVLYARKSLHDEQAR VVLDPNVWTEDGTAAVGATAFTDDGSLMAYTRSDKGSDWMRIFVVRVEDGYVFPDVVKHV KFSSLEWSPDNQGFFYNRYDVPDEDQEDSGTANAQHEFQKVFYHKLGTDQQEDILILSHP EHPKWLTDVTLSWDRRFLIVEISESCSPKNQVWIADLDSIRTPNARDAYDFSKLEWKKIV TNFDAGYEYLTNDGTEFVFLTNKDAPRYKLVKLDILHPEETFWKTLVHETSDLLSSADVV DGNKLILHYFRDVTSQLEVRDLHTGQLLGAIPLPLGSVKQISGERRFPHMLFSFTSFTTP SDIYQMDFAAAVKSGDWTPTLYRQTKVEGADLSQLEVSQAFFDSKDDTKIPMFIMHKKGL KRDGNNPTALYGYGGFNISITPSFTPSWVVAMLNLGMVMAVANIRGGGEYGKAWYEAAIK TKKQVSYDDFQQAAEYLISQQYTSPRRLAIHGGSNGGLLVGACINQRPDLYGAAVLHVGV LDLLRFHKFTIGHAWTSDYGSPENEEEFRSIFQISPLHNIGKARGKGQGHQYPAVFLLTG DHDDRVSPFHSLKFIAELQHSVGSSSKQTNPLVIRVDTNTGHGAGKPVKKSIEEAADVYG FLANVLGIQWHEQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_014330 | 267 S | SESCSPKNQ | 0.997 | unsp | NCLIV_014330 | 267 S | SESCSPKNQ | 0.997 | unsp | NCLIV_014330 | 267 S | SESCSPKNQ | 0.997 | unsp | NCLIV_014330 | 402 S | VKQISGERR | 0.992 | unsp | NCLIV_014330 | 546 S | KKQVSYDDF | 0.997 | unsp | NCLIV_014330 | 564 S | QQYTSPRRL | 0.996 | unsp | NCLIV_014330 | 621 S | SDYGSPENE | 0.998 | unsp | NCLIV_014330 | 711 S | PVKKSIEEA | 0.997 | unsp | NCLIV_014330 | 23 T | RRDETVVET | 0.991 | unsp | NCLIV_014330 | 113 S | YARKSLHDE | 0.995 | unsp |