

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
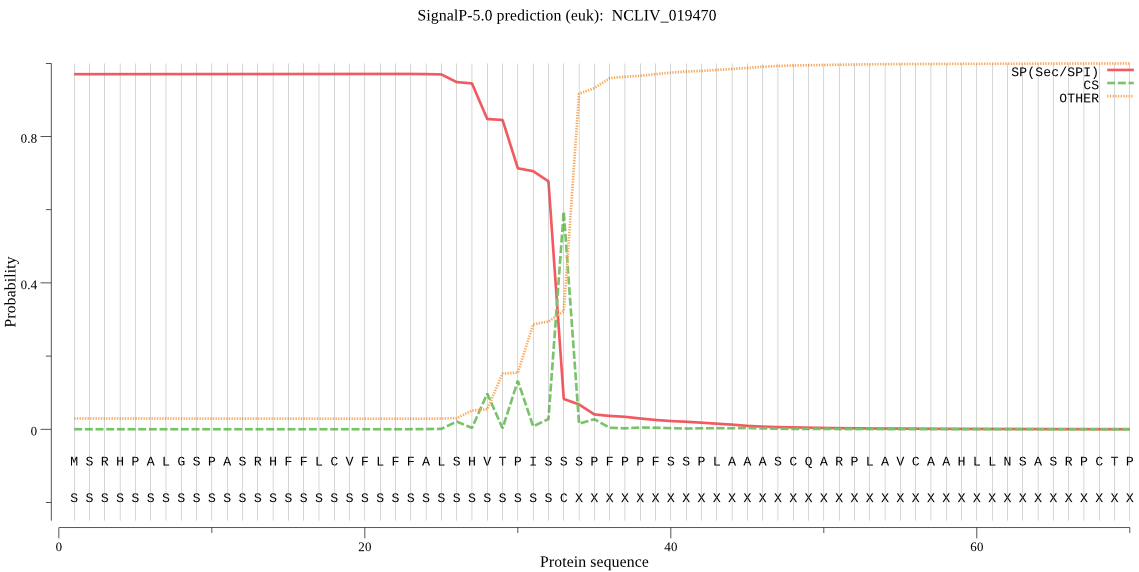
| NCLIV_019470 | SP | 0.153965 | 0.845760 | 0.000275 | CS pos: 33-34. ISS-SP. Pr: 0.8761 |
MSRHPALGSPASRHFFLCVFLFFALSHVTPISSSPFPPFSSPLAAASCQARPLAVCAAHL LNSASRPCTPVQRWSIRESGRSSSSSARSSSSSALSDTPPSLGARLVSFSRKQLHGGSLP CARAVSVRWFPQAKLLPLCTLLHLHCLTTLLSFTALGLNALLSSQHLLLLPSLPFFWRRL LDKNNQLHAFIRGVITPDLPAVQATLTGPFFEELQFRFLLQKCLLSPLLSLVSFWLSVEG KRPLTFFRERPSPFLSADNGGEAKSRGRHAQATRQSYHCSPQRQETSRNRKHSTGTGAEH NNQSVDRARVSQRLGNALRRSRFSSEGERGAHKQKEVKTDTERLCASTKQEQFQGAQEPS PPSPCRVEERLRIAISSLLFAIAHYVPPSSSELDHLESRPPTSHIATPSHFQEPSGPRIS STSPLGSPSLSARTPHRSGLLSAPVDTRSVHGPFPSHRWRIADCIAANRILTAAVQGCVW GFAMERGGLASAVLLHVLHNLQQFALLSAFRWCVFRKQREAKKGSTP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_019470 | 86 S | SSSSSARSS | 0.996 | unsp | NCLIV_019470 | 86 S | SSSSSARSS | 0.996 | unsp | NCLIV_019470 | 86 S | SSSSSARSS | 0.996 | unsp | NCLIV_019470 | 89 S | SSARSSSSS | 0.991 | unsp | NCLIV_019470 | 90 S | SARSSSSSA | 0.994 | unsp | NCLIV_019470 | 293 S | NRKHSTGTG | 0.997 | unsp | NCLIV_019470 | 324 S | RSRFSSEGE | 0.998 | unsp | NCLIV_019470 | 325 S | SRFSSEGER | 0.995 | unsp | NCLIV_019470 | 403 S | RPPTSHIAT | 0.99 | unsp | NCLIV_019470 | 423 S | ISSTSPLGS | 0.993 | unsp | NCLIV_019470 | 75 S | VQRWSIRES | 0.996 | unsp | NCLIV_019470 | 83 S | SGRSSSSSA | 0.996 | unsp |