

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
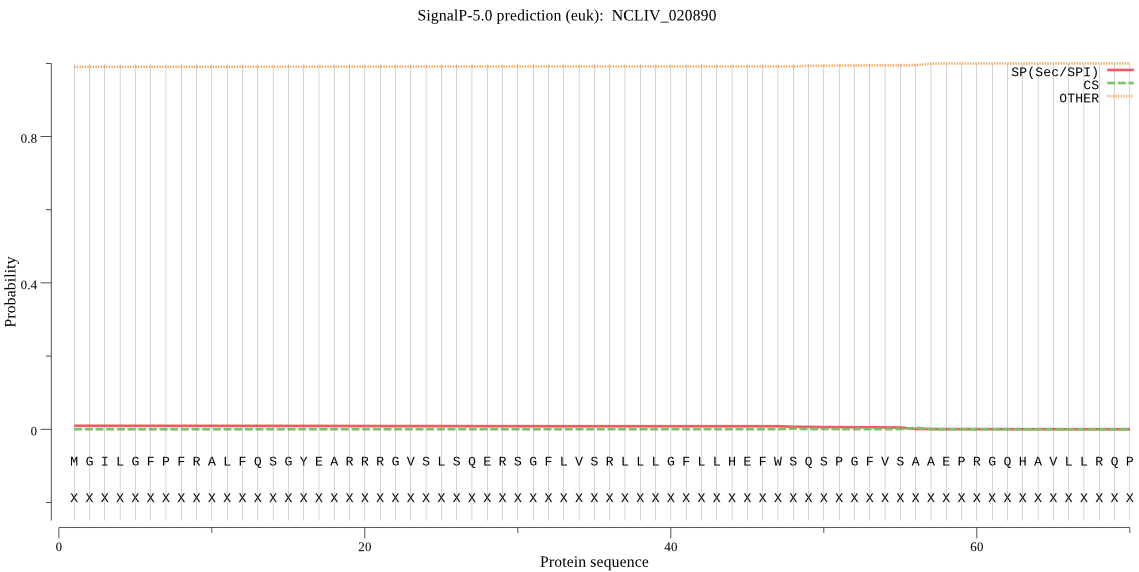
| NCLIV_020890 | OTHER | 0.949767 | 0.003163 | 0.047071 |
MGILGFPFRALFQSGYEARRRGVSLSQERSGFLVSRLLLGFLLHEFWSQSPGFVSAAEPR GQHAVLLRQPFYAEQAEHVRFPMRVGDLTEDNIEALETISQQIAREVHAQQLRRKRERGK KIRPSSSSQQNTPKPAAATEPGKPKAGNKRHPDVKHEKGINEKNRNVDQEATGETEENRE KGETGEKEGNVPKEETEESGKNLDNEETEEKEEIQYSEETEGKGETVGGPVTQDEAGNSE RRNEVRHEDENKPSPSSGGTGETGASGETPLQESEEELESSSTSPEGDVSSLGGRLIIGL TEITFPMETNQSRSETSRAEATDNKYGTAVRDSGHDEAQISGFSHTPASSHSRSETDNKR GAETAYVEEAISRNNVLGHADAVREAKARLQSDRLAELGIMKEMKVLENVGLVVLELNDD LSEDSIRETIKTLWQRNPSTWLIESDTEVTFRGREAFAAGDLVVVPDVDGERGALEDKSN EDIPSRGDKSAEENPENETKADPETHSGVHEEVGQGAAPDDQRRKRKIAEAESDASSSFA RKVTEEGKRRVDEQEATNTRRRGDQERSRTLPSFGEGEKKEARSESNRDGERKRSQVLVS ISAKEAKTGTEHGHSSDEPSDPTPSQHRKQADFSSASHAPTKGERDHVAASAHREADSGG IHISGVVEDFEGTNEARDMPKAHPAGMLKPSTGIIPLSLYEAVDSAVRESLLDNFSSASP LFRSEVLCPSHPVPSPDSCPQRSAQGSRSVDPSPRLPQLASSQPESPSPSRAPLLKGRNS IPLLSGPVRHLGKMPNDSLFSRQWAYHEPRVNVRAVEAWNTVYTHRLSRAANANADGEQV AFGRRLAEHEAPHREIADEEKQRRYVRREKIAKESEVETPSGNRKGAKKEPIIVAVIDTG VDYNHEDLRTQMWRNEKEIPNNGIDDDGNGYVDDVRGYDFEGKTNDPMDLNGHGTHVAGI IAAAANNRRGIAGVNWEVKVMPLKFISRSSAAAEAIDYSLRMGAKISTNSWGYTTPSEGL RLAIERTAKRGQLFVAAVDNAGKDNSVENDFPPNWGYDTRTGAGLKSLLRVANLSPGGIV ASSSNWSPYTVDVAAPGTDIISTIPTGRFPEGYGYKTGTSMATPLVAGVANAYAAVLDAL GDPLPASALQPDASQPPSPAQAVLAFLGGNHGSGSLPSSDSSPASGTSASSSASLPLSPL GGEGGSYALPGLDTLVASMAQLLNPFSRLLTLS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_020890 | 125 S | KIRPSSSSQ | 0.992 | unsp | NCLIV_020890 | 125 S | KIRPSSSSQ | 0.992 | unsp | NCLIV_020890 | 125 S | KIRPSSSSQ | 0.992 | unsp | NCLIV_020890 | 199 S | ETEESGKNL | 0.993 | unsp | NCLIV_020890 | 239 S | EAGNSERRN | 0.992 | unsp | NCLIV_020890 | 274 S | PLQESEEEL | 0.997 | unsp | NCLIV_020890 | 284 S | SSSTSPEGD | 0.997 | unsp | NCLIV_020890 | 312 S | ETNQSRSET | 0.99 | unsp | NCLIV_020890 | 333 S | AVRDSGHDE | 0.998 | unsp | NCLIV_020890 | 352 S | ASSHSRSET | 0.998 | unsp | NCLIV_020890 | 354 S | SHSRSETDN | 0.997 | unsp | NCLIV_020890 | 425 S | LSEDSIRET | 0.996 | unsp | NCLIV_020890 | 479 S | LEDKSNEDI | 0.996 | unsp | NCLIV_020890 | 490 S | RGDKSAEEN | 0.997 | unsp | NCLIV_020890 | 584 S | KEARSESNR | 0.993 | unsp | NCLIV_020890 | 586 S | ARSESNRDG | 0.998 | unsp | NCLIV_020890 | 602 S | LVSISAKEA | 0.992 | unsp | NCLIV_020890 | 615 S | EHGHSSDEP | 0.996 | unsp | NCLIV_020890 | 710 S | AVRESLLDN | 0.996 | unsp | NCLIV_020890 | 762 S | QLASSQPES | 0.993 | unsp | NCLIV_020890 | 766 S | SQPESPSPS | 0.996 | unsp | NCLIV_020890 | 875 S | IAKESEVET | 0.995 | unsp | NCLIV_020890 | 24 S | RRGVSLSQE | 0.994 | unsp | NCLIV_020890 | 26 S | GVSLSQERS | 0.994 | unsp |