

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
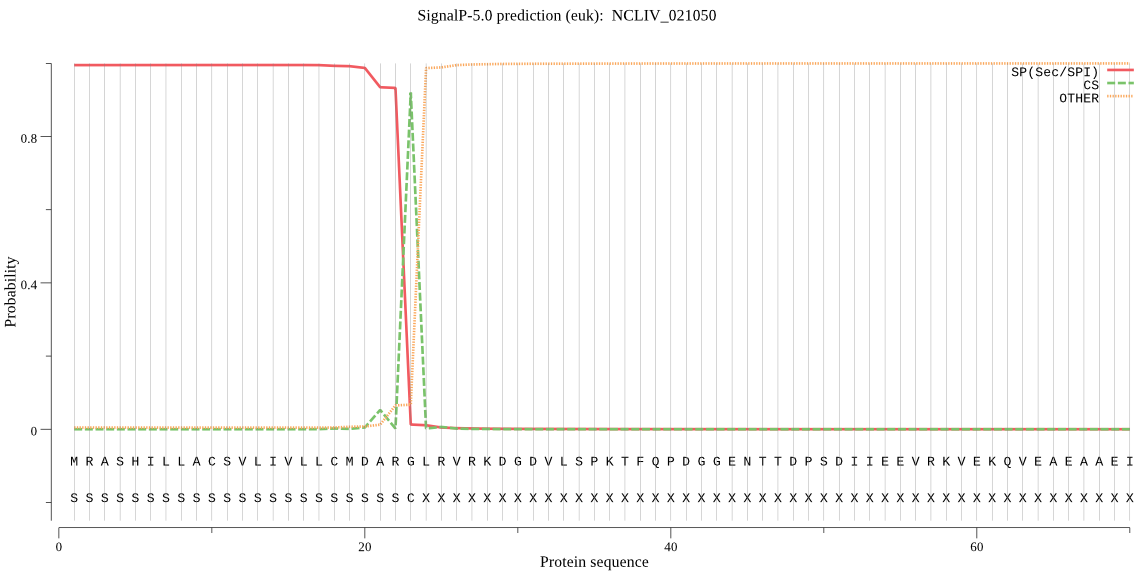
| NCLIV_021050 | SP | 0.005300 | 0.994657 | 0.000044 | CS pos: 23-24. ARG-LR. Pr: 0.8627 |
MRASHILLACSVLIVLLCMDARGLRVRKDGDVLSPKTFQPDGGENTTDPSDIIEEVRKVE KQVEAEAAEIIKAREEHRGFNTLDDGVAPETEGGHGLHASETTPVAELEPQDPDRSLKYP VRLLIVDKPAGDEEETRPSFVQTALHSELAQRVVKELNGHVDVLDESGVVLVDLPANTTD KQLKEVIETAKAQGAIVEPDHMVSAVHTSSRESNDPLLHELWALDPLNMRAAWDILTTAE LGGDRRPLVCVVDTGIDYEHPDLRENMEVNQVELHGKPGIDDDNNGEIDDIYGANMVSDS TDPADDHSHGTHVAGTIGARGDNGVGIAGIAWAPRLIACKFLNARGRGFDSDALRCINYC AKRGADIMNHSWSGSDASEALRQAIEQTAQQGIIHIAAAGNSGRDVDVTPNYPAALSTAV EGLITVGNMKMEKQRDGSKHFSLAESSNYGTKSVQIALPGTDIYSTIPVQERPDDPYGWK TGTSMAAPALSGIVALMLAANPGLSATQIRSILMQSVNRTPELSTRVTWGAMPDAKRCLD AALVTPPEGRRPGNPPSHPPPEASPPESSPPDRQHPHPHPPRPNPPEASPPEPSPPNWQH PHPHPPRPNPPEASPPEPSPPNWQHPHPHPPRPNPPGASPPESSPPNWQHPHPHPPRPNP PEASPPQSSPPEPQRPFSQWPHTPHFFHYHPYPGYNLPYFTYHQSPLPYGPYGRDPCPCA SHPYPADDSPLGSYAPDPSPPQSYPPEPSPSKPSPPEGSSPRVPSPHRHPSRSRLPSAVE PSPPPASPQPSPHPSPPDTSPTKPSTPPPSPSQDPEGRREPSEEDDHKSLSDKSTSHSSE GHAGATPLARVGVLAVFLTVVGLIV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_021050 | 438 S | QRDGSKHFS | 0.991 | unsp | NCLIV_021050 | 438 S | QRDGSKHFS | 0.991 | unsp | NCLIV_021050 | 438 S | QRDGSKHFS | 0.991 | unsp | NCLIV_021050 | 754 S | PSKPSPPEG | 0.996 | unsp | NCLIV_021050 | 765 S | PRVPSPHRH | 0.997 | unsp | NCLIV_021050 | 777 S | SRLPSAVEP | 0.996 | unsp | NCLIV_021050 | 795 S | SPHPSPPDT | 0.996 | unsp | NCLIV_021050 | 810 S | TPPPSPSQD | 0.993 | unsp | NCLIV_021050 | 822 S | RREPSEEDD | 0.998 | unsp | NCLIV_021050 | 831 S | HKSLSDKST | 0.996 | unsp | NCLIV_021050 | 838 S | STSHSSEGH | 0.996 | unsp | NCLIV_021050 | 213 S | SSRESNDPL | 0.993 | unsp | NCLIV_021050 | 373 S | NHSWSGSDA | 0.995 | unsp |