

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_022010 | SP | 0.072855 | 0.907662 | 0.019483 | CS pos: 22-23. ASA-KL. Pr: 0.5835 |
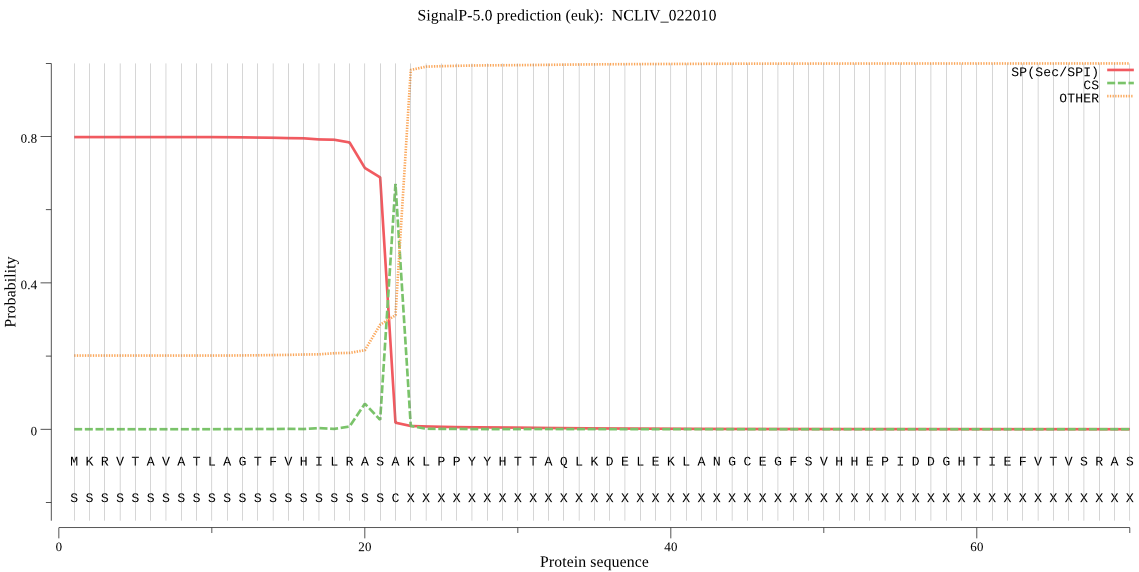
MKRVTAVATLAGTFVHILRASAKLPPYYHTTAQLKDELEKLANGCEGFSVHHEPIDDGHT IEFVTVSRASNNPDRFFMLAGEHPRELISAESALHFLQVLCGKETKLADKARKVLQHTSF QVVVNGNPISRAKVEAGDYCLRGNENNVDLNRNWADHWASAVTSAETNPGNRAFSEPETQ AFAKAVTSFRPDIFLSVHSGEQSLVSVWNEAWLTVC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_022010 | 70 S | VSRASNNPD | 0.991 | unsp | NCLIV_022010 | 175 S | NRAFSEPET | 0.997 | unsp |