

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
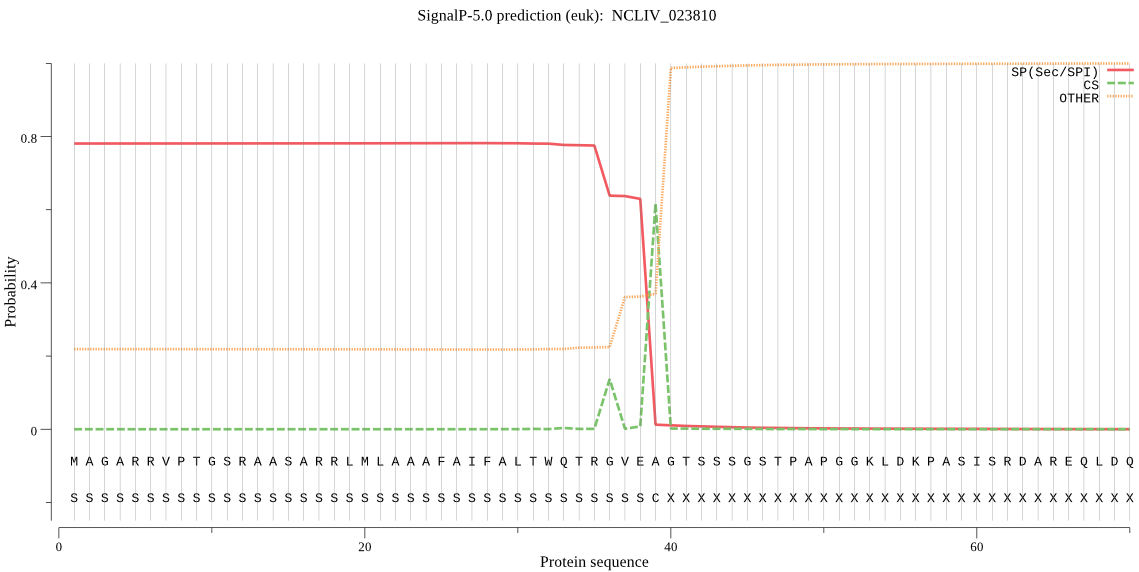
| NCLIV_023810 | SP | 0.049469 | 0.918463 | 0.032068 | CS pos: 39-40. VEA-GT. Pr: 0.5629 |
MAGARRVPTGSRAASARRLMLAAAFAIFALTWQTRGVEAGTSSSGSTPAPGGKLDKPASI SRDAREQLDQAEVLAMYMHHRHITPVHFLTVLLEVQGDYSITKSLQECGGNLYELKKALS NVLIKLPTSSEVYIVDEVESTAQLKRVALRALNIAQKDKKQTIGMEHLILSLFEERNLRT MLEQVGCNVKVFLEHSTAMYETKFKEDVANRSKKKQGSTSASSTSESTPENSDEEFVKSF GVDMTQLAADGKLEPVVGRNKEIKEVITVLSRKGKGNPCLIGEPGVGKTAVVEGLAQRLI EGMVPKSLENKILFAVDLGALIAGATYRGEFEKRMKALLRYAVGQEGRVILFIDEIHMLM GAGKSDGTVDAANLLKPPMARGEIRLVGATTQEEYKIIERDAAMERRLKPILVEEPTTDR AIYILRKLKEKFERHHEMTISDEAIVSAVMLSHKYIKNRKLPDKAIDLLDEAAATKRVKW DLRTVDDDEMASEKKQKEKLEEEHRKVEEQDKKRIEGQHHGAGTEEELKLLEKEIEQEAL EEEQELVLTADDVAVVISSWTGIPVAKLTDDEKGKILRLSDLLHSRVIGQEDAVKAVADA MVRARAGLSREGMPVGSFLFLGPTGVGKTELAKALAAEMFHSEKNLIRIDMSEFSEAHSV SRLIGSPPGYVGHEAGGQLTEAVRRRPHSVVLFDEIEKGHQQILNIMLQMLDEGRLTDGK GLLVDFTNCVIILTSNVGAQYIISAYEQGEKDSRSALATFDPKSGSLEQFAEATKAAQVA AESDDAASAKTDKKGAVLDKNKKSGNWKKEARRKILSEIAGSGLLKPEVVNRFSGVIIFE PMKPSDVRKILSIQSKDLKEALARKGIELVIDGSAEEFIVQRSYTHKFGARPLKRFVDNQ IGAKMAPWILSGYLQPGMRVTVVASKRNKNTLEAHVCKVVMGKCDRTTRKARVLASVAKK DDSSPDDPGR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_023810 | 227 S | STSESTPEN | 0.991 | unsp | NCLIV_023810 | 227 S | STSESTPEN | 0.991 | unsp | NCLIV_023810 | 227 S | STSESTPEN | 0.991 | unsp | NCLIV_023810 | 232 S | TPENSDEEF | 0.995 | unsp | NCLIV_023810 | 492 S | DEMASEKKQ | 0.993 | unsp | NCLIV_023810 | 659 S | SEAHSVSRL | 0.992 | unsp | NCLIV_023810 | 689 S | RRPHSVVLF | 0.994 | unsp | NCLIV_023810 | 788 S | DDAASAKTD | 0.99 | unsp | NCLIV_023810 | 963 S | KKDDSSPDD | 0.997 | unsp | NCLIV_023810 | 964 S | KDDSSPDDP | 0.998 | unsp | NCLIV_023810 | 15 S | SRAASARRL | 0.995 | unsp | NCLIV_023810 | 223 S | TSASSTSES | 0.997 | unsp |