

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_024640 | OTHER | 0.999921 | 0.000004 | 0.000075 |
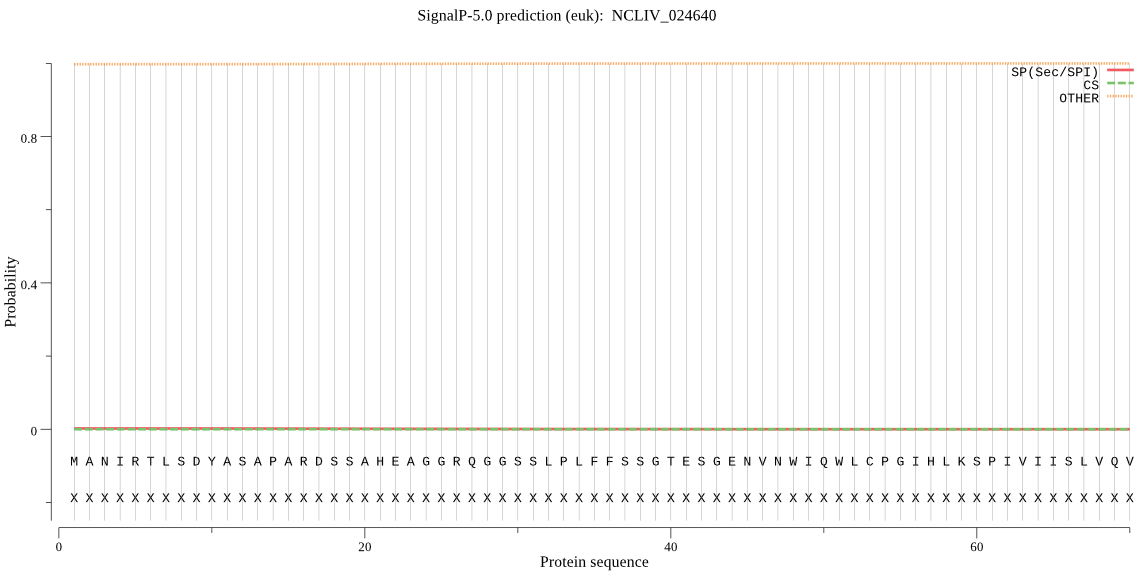
MANIRTLSDYASAPARDSSAHEAGGRQGGSSLPLFFSSGTESGENVNWIQWLCPGIHLKS PIVIISLVQVAVYIASLALGLAPNEVLAPTPQTLAMFGANIPELIRIGQVWRLICPLFLH LNLFHILMNLWVQSRVGLTMEEKYGSKKLLATYFGVGILANMISAAVLFCGQMKAGASTA VFALIGVQLAELALIWHALQDRNSAILSVCICLFFVFVSSFGGHTDGIGHLGGLVMGFAA GIWLNENSDVKPTWYDRGLVTSKVALAIAPIVSSIFIFLVPRC
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_024640 | 19 S | ARDSSAHEA | 0.994 | unsp |