

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
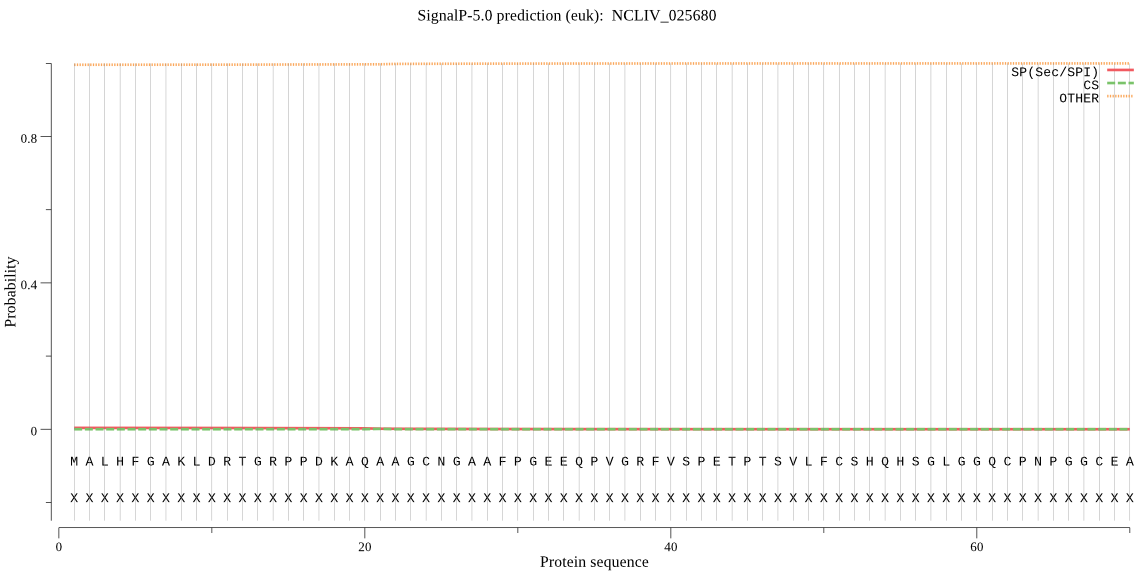
| NCLIV_025680 | OTHER | 0.998526 | 0.001405 | 0.000069 |
MALHFGAKLDRTGRPPDKAQAAGCNGAAFPGEEQPVGRFVSPETPTSVLFCSHQHSGLGG QCPNPGGCEAPSCLQGATACEERNGDTSASPASNCVSNSGGNAHGLDGEGVPCSAAESRR SGKVGGLGGEKAGSLGKESPARQKRVLSEAEINLEKILEEDSGRDCNADYPVPKWLRGQG VCSPFTYFRSFKASLGIINYDLRGPPGGPIVVTFHGLNGTQLTFYDMQEVLARFGYRTLI FDLYGHGLSASPRYSFFLKRYGLQFFIKQTDELLEHLGLQNERISVVGFSMGCVIAAEYA LHRKELVDRICLVAPAGLLPNKPFAVRVLQRFGCKKGFVQKYEEEEHEERRRSSASAAAA GPTTAEAPEEPRKKSLSWRKRPSEGDRKERRREAGRASGKAFSCRQGSAAADSTGAPGDA KRPSEQGAVSGVSPGELLWRRLMWQLYVKKGVVATFVGCVTHVPLWDARKIYERVGETGK PVLMLWGQDDAVAPLSCATSLRALMPNSHLITFPACSHLVLADRAQASIGCVMAFLDFPA TCGMQQWRFALPFDSEGTYVQPQDRAPPGTRAEDYLHELGYSPKFTIRLAQTPDGAGARQ GPLRRRTREETDTDQQLGKRSDAPRSDGKPDCRSDRRSTCEAEGGERAIQRRVRAQMKAE LAAHERGSSVARAKTSAGSLEERQQRRIQGPKRSRDCGLAPVGERADDETPGTEEEDAFG ETRLLPVERFTDSPVEGENGASEDSEFEEAVLMLPDAGDEETAGGKQERTSETVHLPLDG LRSRSASAAAAAATSHLLSFDSNSTQEILLQGSGDQVLVPTLSRVSP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_025680 | 354 S | RRRSSASAA | 0.997 | unsp | NCLIV_025680 | 354 S | RRRSSASAA | 0.997 | unsp | NCLIV_025680 | 354 S | RRRSSASAA | 0.997 | unsp | NCLIV_025680 | 377 S | KKSLSWRKR | 0.996 | unsp | NCLIV_025680 | 383 S | RKRPSEGDR | 0.997 | unsp | NCLIV_025680 | 398 S | AGRASGKAF | 0.994 | unsp | NCLIV_025680 | 433 S | VSGVSPGEL | 0.995 | unsp | NCLIV_025680 | 638 S | SDRRSTCEA | 0.998 | unsp | NCLIV_025680 | 679 S | TSAGSLEER | 0.994 | unsp | NCLIV_025680 | 733 S | RFTDSPVEG | 0.996 | unsp | NCLIV_025680 | 771 S | QERTSETVH | 0.995 | unsp | NCLIV_025680 | 826 S | LSRVSP--- | 0.995 | unsp | NCLIV_025680 | 121 S | ESRRSGKVG | 0.997 | unsp | NCLIV_025680 | 353 S | ERRRSSASA | 0.995 | unsp |