

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
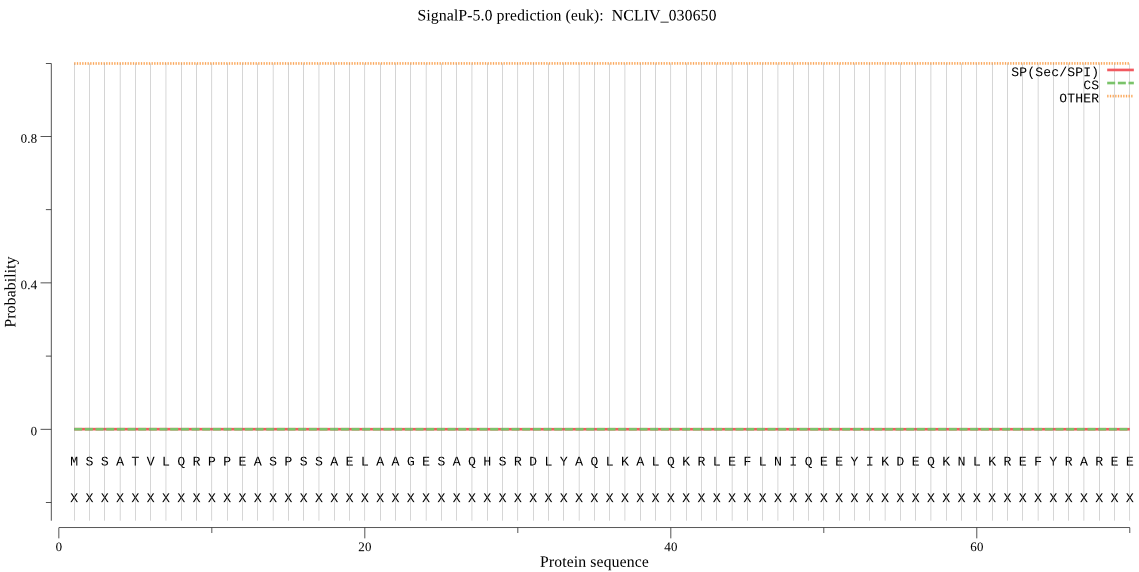
| NCLIV_030650 | OTHER | 0.999979 | 0.000007 | 0.000013 |
MSSATVLQRPPEASPSSAELAAGESAQHSRDLYAQLKALQKRLEFLNIQEEYIKDEQKNL KREFYRAREELKRIQSVPLVIGQFMELINANEGIVASTAGSSYVVRILSTLNRQERRIEL LKTGCSVALHRHSHSVVDILPPEADSTIQTLQMQEKPDVTYSDIGGMDIQKQEIREAVEL PLTCPELYQQIGIDPPTGVLLYGPPGTGKTMLAKAVANNTTATFIRVVGSEFVQKYLGEG PRMVRDVFRLARENSPAIVFIDEVDAIATKRFDAQTGADREVQRILLELLNQMDGFDQTT TVKVIMATNRADTLDPALLRPGRLDRKIEFPLPDRRQRRLIFQTITAKMNLSDEVDLEDY VSRPEKVSAADIAAICQEAGMQAVRKNRYVILPKDFEKGWKAHVRKHERDFDFYSF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_030650 | 14 S | PPEASPSSA | 0.99 | unsp |