

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
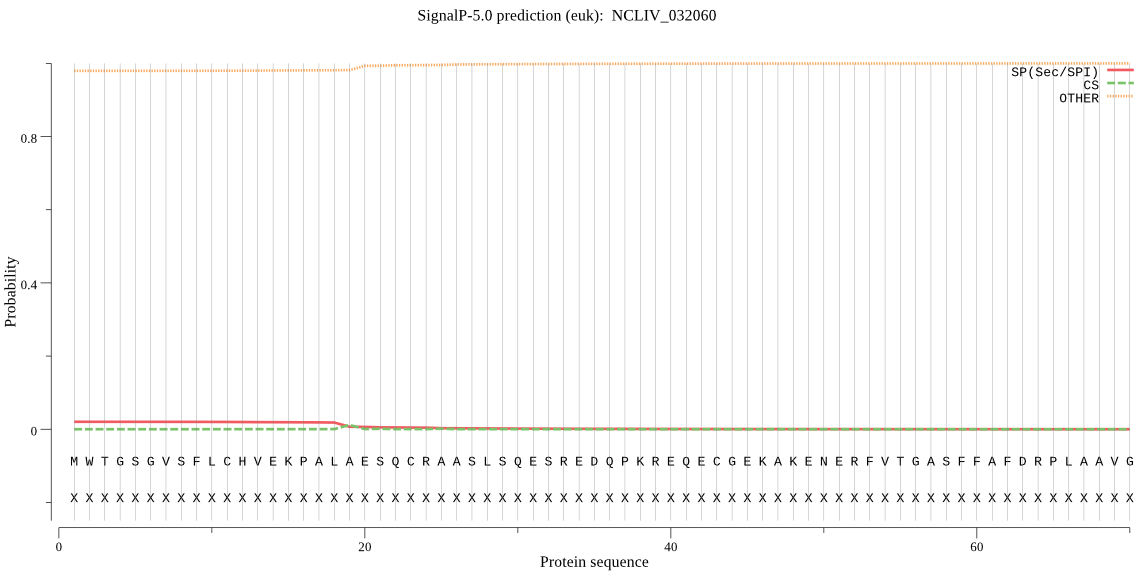
| NCLIV_032060 | OTHER | 0.950100 | 0.029811 | 0.020090 |
MWTGSGVSFLCHVEKPALAESQCRAASLSQESREDQPKREQECGEKAKENERFVTGASFF AFDRPLAAVGRLFGSAPATRDFGAVQIGNSVQCIHGILFGGGSVFGLDAVGGLLEFQRQR GIGVSTIAGPIPVCPALGIFDVPALPPGPSSSETRAFALPQFVTSADVSTACAEAVRVQA SVPASLPRSDREGDKHREAVEKEGTPALADSPLRVRCRLGSNGTVSTAFSHAWTRLSLRK DADRTPAASTSPGQSTASSGASGSPSLEETASWIGSIGVGVGATCARVGECAGFAPRKGG WGEACRQRPWRGERREGKAGDTERGSAAEAGLETLKVQAFIVVNSYGDIVAAGKGSEGSV VAGPLKDGVTYAAADLLQEAGIHVSLNRDALWRVAAMAGAGMARAIHPIFSPVDGDAVIA VSTGELTHGDTDPTLSEIIVGTMAAECVAEAIQQAAAASNNL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_032060 | 266 S | SGSPSLEET | 0.997 | unsp | NCLIV_032060 | 266 S | SGSPSLEET | 0.997 | unsp | NCLIV_032060 | 266 S | SGSPSLEET | 0.997 | unsp | NCLIV_032060 | 326 S | TERGSAAEA | 0.993 | unsp | NCLIV_032060 | 411 S | HPIFSPVDG | 0.996 | unsp | NCLIV_032060 | 32 S | LSQESREDQ | 0.998 | unsp | NCLIV_032060 | 189 S | SLPRSDREG | 0.997 | unsp |