

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
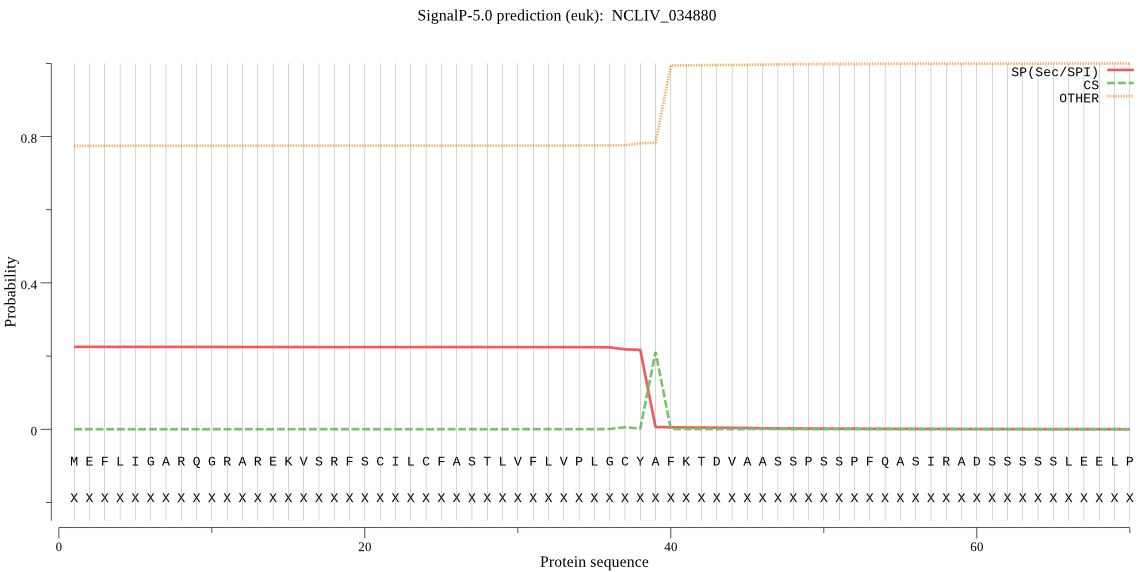
| NCLIV_034880 | OTHER | 0.525494 | 0.474225 | 0.000281 |
MEFLIGARQGRAREKVSRFSCILCFASTLVFLVPLGCYAFKTDVAASSPSSPFQASIRAD SSSSSLEELPVSVQSTLPSPSSAPEGVNRGVVEKTSSRKIDSSPAARESAASSSPHPSLF FSLSVPASSRSDSGDTTFAVSGERGAADEEPQEASMSSTAGPNSDVDHALFVVPGIAGSG LFATVTNASFEACGKSPINYAVPFRVWASLSLLLPPVTHQRCWVEMMKMTVDENGETYTA QEGVHVEVDGYGGIHAIDYLDYYMNNTYGVPASAYMHGMLRTLLSLHYAQFVTLRGVPYD WRLPPWQLNYAQLKADIEDRYTELNNRKVDLIAHSLGSIILCYFLNRVVDQAWKDKYIGS MTLVAAATGGSFKAVKSLLSGYDDGTDIDIWNVIDFSLFPAVLLRDLLQTMGSIYALLPD PAVYGRDHVVVRVARPPTRVPALSSAAASPSQSPAGRTAGGASHVAERAKDPISNLSASS GEMRRRLDAVVTDEAAGQGSRARDRESEQSFSRVSDLARDSLDVEKELAEGETNTRVDRS PVADSVGEHMEEEALAKAEERREALLREFQLQNRLSHEDRAARQRERYSLFVEKEKRRLD EALRVAKEEGLEEDVYTLSNWTSLLPADLQRRVKTAQDMMAGVVADPGVPVRCIWSKFTQ PTTDVAYYYASGSFDSHPIPIFDYGDNTVPLPSLSLCASWRSTVQVKAFDNLDHMFLFAD RGFNSYIYDLFTPNPQQSATALSTDEGKATKHDGFPAAQESRRGTVAATEDRA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_034880 | 65 S | SSSSSLEEL | 0.997 | unsp | NCLIV_034880 | 65 S | SSSSSLEEL | 0.997 | unsp | NCLIV_034880 | 65 S | SSSSSLEEL | 0.997 | unsp | NCLIV_034880 | 112 S | ESAASSSPH | 0.99 | unsp | NCLIV_034880 | 129 S | VPASSRSDS | 0.996 | unsp | NCLIV_034880 | 133 S | SRSDSGDTT | 0.996 | unsp | NCLIV_034880 | 141 S | TFAVSGERG | 0.996 | unsp | NCLIV_034880 | 164 S | AGPNSDVDH | 0.995 | unsp | NCLIV_034880 | 380 S | KSLLSGYDD | 0.991 | unsp | NCLIV_034880 | 449 S | SAAASPSQS | 0.993 | unsp | NCLIV_034880 | 479 S | NLSASSGEM | 0.991 | unsp | NCLIV_034880 | 540 S | RVDRSPVAD | 0.991 | unsp | NCLIV_034880 | 545 S | PVADSVGEH | 0.993 | unsp | NCLIV_034880 | 576 S | QNRLSHEDR | 0.997 | unsp | NCLIV_034880 | 743 S | ATALSTDEG | 0.993 | unsp | NCLIV_034880 | 48 S | VAASSPSSP | 0.991 | unsp | NCLIV_034880 | 61 S | IRADSSSSS | 0.994 | unsp |