

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
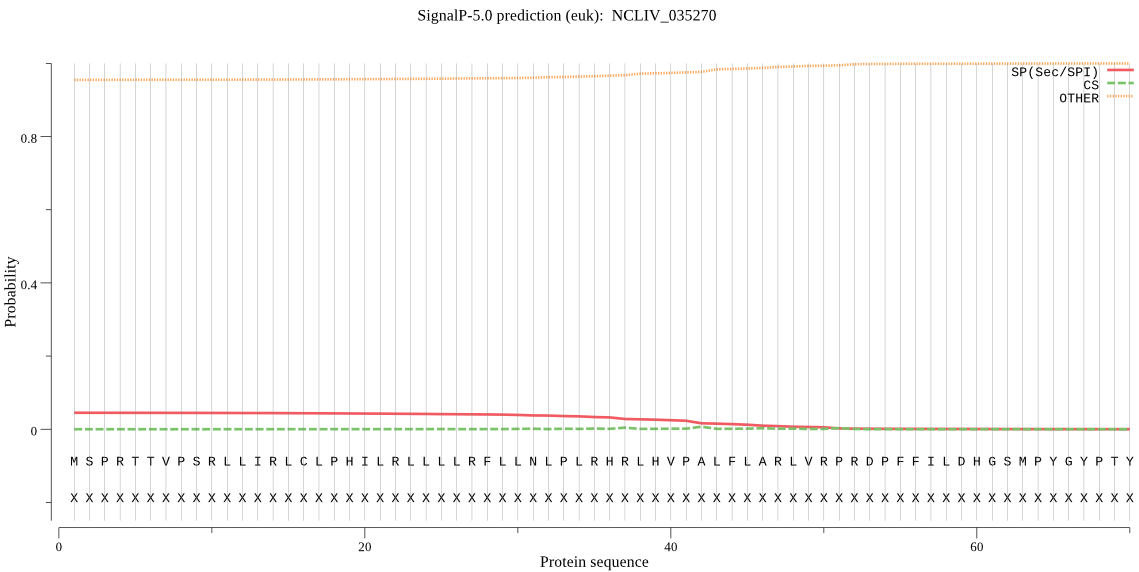
| NCLIV_035270 | OTHER | 0.903691 | 0.054906 | 0.041403 |
MSPRTTVPSRLLIRLCLPHILRLLLLRFLLNLPLRHRLHVPALFLARLVRPRDPFFILDH GSMPYGYPTYDEAFLMMAQLLKTYGPSLISEHRIGHSTEGRPIVAYRVGGTSKPRAAAAP VSPATPASAPAGTSSLRGTKSEPLREQGTSEGGTQPDNLHGKKDEFPEILITGLHHAREP MSMTMCIYFIARLLRDYQAGEPEAVYLVHMREFWVIPLLNPDAYVAIETTGNMALRKNRR RFSSESRPPNAQLEDEGVDLNRNYAFNFLADQPEGSDDFGGPFPFSEPETAAVKFLVEQY RHADRALPQSSASPAAGSRMVDKHEHSDAVRAPANAQAPLLSPPLHGVRDFSPPQLSPFL RLLGRFEVALNFHTYGEVWTRPFNCCKNMPLPLWAQKAFEELKVALAVPTLNSAPNIPVL GYPTFGEADDWLLHAHSVLSMSPEIGWEEGGFWQTVGEQRESLRLNFPRIVTAAIKAGSE VGVRLALGQGLSAPHGARLPAIGTGKAKRVGGGHARTDDRRRAPSRTEGEADAEEGAEEA KDARTAEASSVLDSVVPRDRLTFVENVLRQKGWASILKQDDAGVSVHTIEVVNTGLLPLQ GPATLLFLTGLPFFSSPVAASLFSSVDAAAAASLPPIRSTLERRFTGSRSKTKLPPPHAL RTLQAFRSYQSPSYTPSSSSSSSSSSSSSSSSSSRSAVPSSADLRFDVLDEGGDGVGEKQ AWNFVAFDEEWRTPLSVSFFSPSSLSRKAEAPHGPELPGLLVELPARLRGRDLESGDAGV LRVHLLETTGTSDASSPALSLSERLSSPYQSSNSSFPRSAQSAAVGACVLEAGAPEPEKA VKEVVPERGERQQPWFCRCGATKAEVEGDAPAGEADRDGKWISLPLFLENSKTSLLCAAT LSLLSRHRPVSSPSVSSSREASRGFEAGRRLGGRCKGDTFFWDLDVPLHYANRRSSVSFA LLMLLTLVLVLALALRLRYK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_035270 | 525 S | RRAPSRTEG | 0.998 | unsp | NCLIV_035270 | 525 S | RRAPSRTEG | 0.998 | unsp | NCLIV_035270 | 525 S | RRAPSRTEG | 0.998 | unsp | NCLIV_035270 | 550 S | AEASSVLDS | 0.996 | unsp | NCLIV_035270 | 682 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 683 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 684 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 685 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 686 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 687 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 688 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 689 S | SSSSSSSSS | 0.99 | unsp | NCLIV_035270 | 690 S | SSSSSSSSS | 0.994 | unsp | NCLIV_035270 | 692 S | SSSSSSSRS | 0.994 | unsp | NCLIV_035270 | 693 S | SSSSSSRSA | 0.995 | unsp | NCLIV_035270 | 700 S | SAVPSSADL | 0.991 | unsp | NCLIV_035270 | 746 S | PSSLSRKAE | 0.992 | unsp | NCLIV_035270 | 807 S | ERLSSPYQS | 0.996 | unsp | NCLIV_035270 | 917 S | PSVSSSREA | 0.995 | unsp | NCLIV_035270 | 918 S | SVSSSREAS | 0.995 | unsp | NCLIV_035270 | 956 S | NRRSSVSFA | 0.997 | unsp | NCLIV_035270 | 243 S | RRRFSSESR | 0.997 | unsp | NCLIV_035270 | 244 S | RRFSSESRP | 0.996 | unsp |