

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
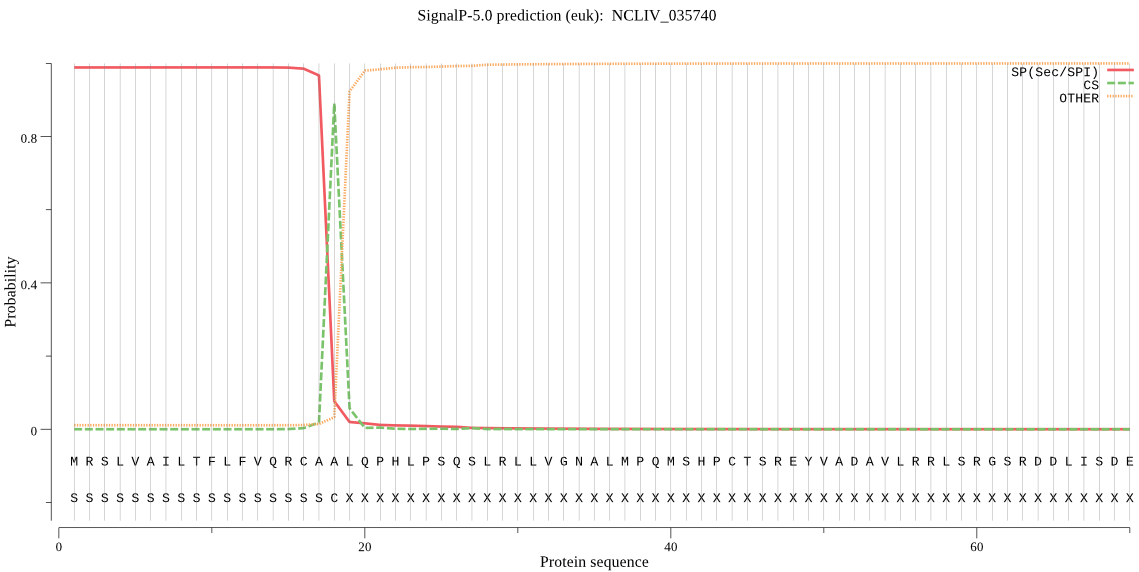
| NCLIV_035740 | SP | 0.019548 | 0.970174 | 0.010277 | CS pos: 18-19. CAA-LQ. Pr: 0.8969 |
MRSLVAILTFLFVQRCAALQPHLPSQSLRLLVGNALMPQMSHPCTSREYVADAVLRRLSR GSRDDLISDESVNNLLPGCLEVRDMPYRMHAREVPGDGACLFASVAACLWWSAFETHADL NDPAFLDMIGSLRQLAVDTLQDTNVSTLVLEGDEVLPRSSLVNLAAADYNTTPAAYCERM RLPNTWGGGPEIVALSHALRRVIVVYEVHDPSLRSGNSQGSSAHCMHPVCLGQLSSSDGT AQNAPNQTALKVIACLGFPQNVAEEPLHILFTSSAACSTGLRPHAGQTADHFLPLFPAVS RK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_035740 | 59 S | LRRLSRGSR | 0.994 | unsp | NCLIV_035740 | 62 S | LSRGSRDDL | 0.997 | unsp |