

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
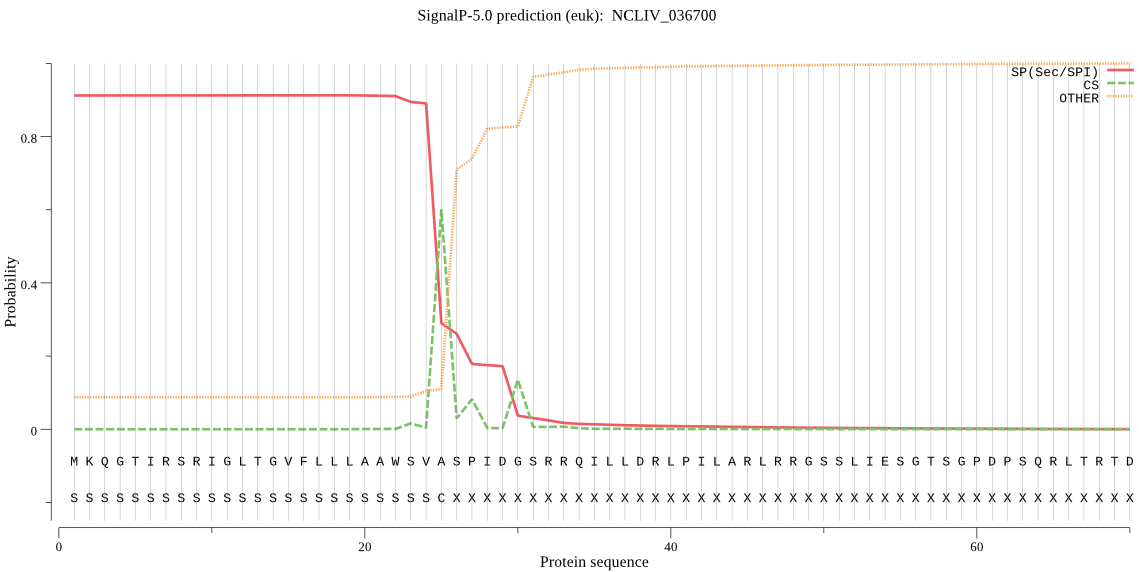
| NCLIV_036700 | SP | 0.014030 | 0.984073 | 0.001897 | CS pos: 30-31. IDG-SR. Pr: 0.5581 |
MKQGTIRSRIGLTGVFLLLAAWSVASPIDGSRRQILLDRLPILARLRRGSSLIESGTSGP DPSQRLTRTDKIRKPRSDRRSYRYVELPNELRALLVSDPECDEAAASMRVGVGSTSDPPD IPGLAHFTEHMLFQGSKRFPGTHDFFDFVHDHGGYTNAFTSKFSTVFSFSIGPQFLEPGL DRLADIFSAPLLKDENLLKEVNAVHSEYIVDLTDDNHRKHHLIRQTASGGPLSNFTVGNL ESLVERTKQQGIDPVKAMRQFHNRWYSSNLMTLAVVGRESLDILESHVRQHFSNVPNGRV TAPVFEECSETFVPLAPNELGTQVNVSLTEEGRSKDSVYKIGLRLFTFLRNLGAARPERW RVTEMAKIRQLGFTFADMPDPYALTVRAVEGLYYYTPEEVMAGDQLIYRFDPTLVQQYLQ KFLVPDNVRLFIFDKKLAAGASREEYWFKIKHMVEPILEPVLKTWKDISKSPLDMVQKIM EMGRMALPAPNRYLPNNVAVLHPPVSAESRRETGTFPEPLVFPEDHACSNKCTVFHKQDT TFGSPKAVVELQMYYTGNHPDDVRERILTTLYVLSIKLALRERFSDAHRGGLSLGLSSGV TLGSPTLPTVKRQKVLTLYAAGFSDKLDYFFRAVAKNLAAAESASDENRTSPSPTALGVT VSHGLRQAVHSKITLPSSVLSSVHSSRLDSTAVRFPGGGGLARRTLLHSQITTLQQKPAS VALTVMRSPITNVGIPVSRSRQGGDTGSAAAGGSFTGEKPLLEKRFVKLALDSLRTQLRV TTFNRSPLRQANDAVLELVMHPYIPVHKVYAAFLEMEKEYRTLDTIFEEVADWGKKIWNE AAIEGLVQGNISSESAAQLVGDVISLLPLKNIVSADSIAKPTISPLSSLGEASSAVSDSS PQRLSVVSDTPTSNDTAYVPSSENQTPASTGHAPSVVFRGPPLAERILKALPPFGKVRAI RRRWEVRGANAHSLQYRPLRFSSAWLNPNPTEERRILLRRVLRRRLAAMRQKAQTDTQQN PFVVFAFRRPPTVARRLRIAQGRSFLQSQGEVSKGSEDPAAGEAGDGPCAEGACAQRLCR GSECQQTPKRASESPCTGEACRGSPCTSLECQRSPSSKAEQPCTDNDCSGPCRGPGCSSA AASQLSDCTEAECGRTCPGCKHLEEKSLHLRSKRKNMNPNDKKNQAYLVIEVRQMEDRRK LVSPVEGRDTLKNIRDRAILHMISQWMSQRFFNRLRTEQQLGYLTAMRYSRLEDKYYYGF FITSTYDPAQVADRIVQFIYAERSKALSQEEFATLQRAAIDVWKQKPKNIFEEFSKNRRQ VILGDRLFDINERMVAELERVTPEEIQSFKERTMFNAPWLLMEVYSQREEPQKLPQASIT SGEGSANPWLDVNPELLSNINVPESEDS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_036700 | 58 S | ESGTSGPDP | 0.996 | unsp | NCLIV_036700 | 58 S | ESGTSGPDP | 0.996 | unsp | NCLIV_036700 | 58 S | ESGTSGPDP | 0.996 | unsp | NCLIV_036700 | 77 S | RKPRSDRRS | 0.997 | unsp | NCLIV_036700 | 81 S | SDRRSYRYV | 0.997 | unsp | NCLIV_036700 | 114 S | VGVGSTSDP | 0.996 | unsp | NCLIV_036700 | 327 S | QVNVSLTEE | 0.99 | unsp | NCLIV_036700 | 471 S | DISKSPLDM | 0.996 | unsp | NCLIV_036700 | 509 S | VSAESRRET | 0.997 | unsp | NCLIV_036700 | 651 S | ENRTSPSPT | 0.997 | unsp | NCLIV_036700 | 1087 T | ECQQTPKRA | 0.991 | unsp | NCLIV_036700 | 1116 S | QRSPSSKAE | 0.991 | unsp | NCLIV_036700 | 1117 S | RSPSSKAEQ | 0.992 | unsp | NCLIV_036700 | 1203 S | RKLVSPVEG | 0.998 | unsp | NCLIV_036700 | 1250 S | AMRYSRLED | 0.992 | unsp | NCLIV_036700 | 1288 S | SKALSQEEF | 0.998 | unsp | NCLIV_036700 | 26 S | WSVASPIDG | 0.995 | unsp | NCLIV_036700 | 51 S | RRGSSLIES | 0.991 | unsp |